
Carnation ringspot virus (isolate Lommel) (CRSV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Regressovirinae; Dianthovirus; Carnation ringspot virus
Average proteome isoelectric point is 8.01
Get precalculated fractions of proteins

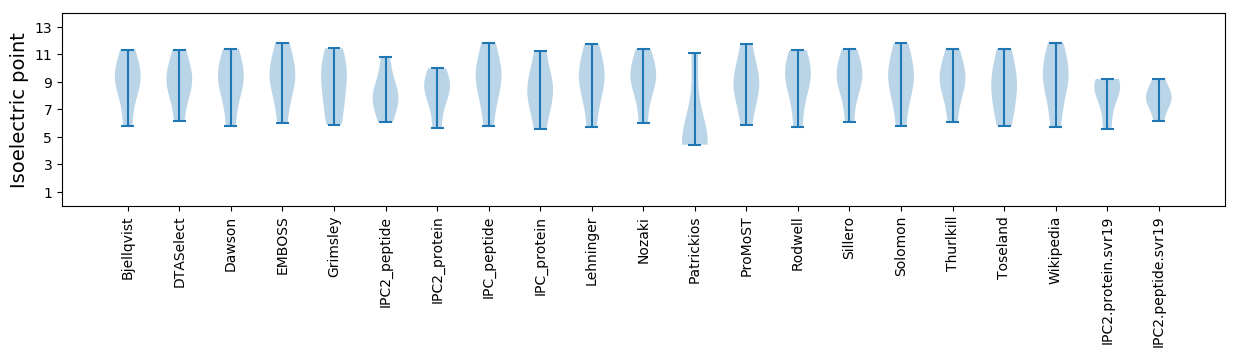
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>sp|Q66096-2|RDRP-2_CRSVL Isoform of Q66096 Isoform protein p27 of RNA-directed RNA polymerase OS=Carnation ringspot virus (isolate Lommel) OX=652597 GN=ORF1 PE=4 SV=2
MM1 pKa = 7.86AIFEE5 pKa = 4.99LFSFDD10 pKa = 4.03IDD12 pKa = 3.99KK13 pKa = 11.23LLVWVSKK20 pKa = 9.79FSPGKK25 pKa = 9.66ILSAICRR32 pKa = 11.84LGLDD36 pKa = 2.89SWNNFRR42 pKa = 11.84KK43 pKa = 9.4WFWGLNFDD51 pKa = 3.5KK52 pKa = 11.23HH53 pKa = 5.07EE54 pKa = 4.07WAVDD58 pKa = 3.09HH59 pKa = 6.77FMPLMPRR66 pKa = 11.84FSSDD70 pKa = 2.55MDD72 pKa = 3.51EE73 pKa = 4.03VAKK76 pKa = 10.78RR77 pKa = 11.84IVTPKK82 pKa = 10.39SKK84 pKa = 10.54PKK86 pKa = 10.92LEE88 pKa = 4.78DD89 pKa = 3.35CLEE92 pKa = 4.08IDD94 pKa = 3.97TAVEE98 pKa = 3.86EE99 pKa = 4.83CFNEE103 pKa = 4.37EE104 pKa = 3.89CFEE107 pKa = 4.11PQEE110 pKa = 4.54DD111 pKa = 3.81GSMKK115 pKa = 10.19LKK117 pKa = 10.41RR118 pKa = 11.84VAAPQQIKK126 pKa = 9.94RR127 pKa = 11.84VRR129 pKa = 11.84TGMIEE134 pKa = 3.95DD135 pKa = 5.31AISAVEE141 pKa = 3.48ARR143 pKa = 11.84IRR145 pKa = 11.84NRR147 pKa = 11.84HH148 pKa = 5.74MIMGDD153 pKa = 3.29DD154 pKa = 3.49MGLVDD159 pKa = 4.55EE160 pKa = 4.8AAVRR164 pKa = 11.84ATAMDD169 pKa = 2.88ICGEE173 pKa = 4.11YY174 pKa = 10.14KK175 pKa = 10.25INEE178 pKa = 3.88HH179 pKa = 5.62HH180 pKa = 6.47TRR182 pKa = 11.84CIIYY186 pKa = 9.64AAAYY190 pKa = 10.02RR191 pKa = 11.84AMTPDD196 pKa = 3.43QEE198 pKa = 5.72SIDD201 pKa = 4.03ATKK204 pKa = 9.72MAYY207 pKa = 9.67NPKK210 pKa = 9.32SQARR214 pKa = 11.84RR215 pKa = 11.84DD216 pKa = 3.6LVSILRR222 pKa = 11.84KK223 pKa = 9.94NISFGGFKK231 pKa = 10.66SLEE234 pKa = 4.18DD235 pKa = 3.89FLNN238 pKa = 3.66
MM1 pKa = 7.86AIFEE5 pKa = 4.99LFSFDD10 pKa = 4.03IDD12 pKa = 3.99KK13 pKa = 11.23LLVWVSKK20 pKa = 9.79FSPGKK25 pKa = 9.66ILSAICRR32 pKa = 11.84LGLDD36 pKa = 2.89SWNNFRR42 pKa = 11.84KK43 pKa = 9.4WFWGLNFDD51 pKa = 3.5KK52 pKa = 11.23HH53 pKa = 5.07EE54 pKa = 4.07WAVDD58 pKa = 3.09HH59 pKa = 6.77FMPLMPRR66 pKa = 11.84FSSDD70 pKa = 2.55MDD72 pKa = 3.51EE73 pKa = 4.03VAKK76 pKa = 10.78RR77 pKa = 11.84IVTPKK82 pKa = 10.39SKK84 pKa = 10.54PKK86 pKa = 10.92LEE88 pKa = 4.78DD89 pKa = 3.35CLEE92 pKa = 4.08IDD94 pKa = 3.97TAVEE98 pKa = 3.86EE99 pKa = 4.83CFNEE103 pKa = 4.37EE104 pKa = 3.89CFEE107 pKa = 4.11PQEE110 pKa = 4.54DD111 pKa = 3.81GSMKK115 pKa = 10.19LKK117 pKa = 10.41RR118 pKa = 11.84VAAPQQIKK126 pKa = 9.94RR127 pKa = 11.84VRR129 pKa = 11.84TGMIEE134 pKa = 3.95DD135 pKa = 5.31AISAVEE141 pKa = 3.48ARR143 pKa = 11.84IRR145 pKa = 11.84NRR147 pKa = 11.84HH148 pKa = 5.74MIMGDD153 pKa = 3.29DD154 pKa = 3.49MGLVDD159 pKa = 4.55EE160 pKa = 4.8AAVRR164 pKa = 11.84ATAMDD169 pKa = 2.88ICGEE173 pKa = 4.11YY174 pKa = 10.14KK175 pKa = 10.25INEE178 pKa = 3.88HH179 pKa = 5.62HH180 pKa = 6.47TRR182 pKa = 11.84CIIYY186 pKa = 9.64AAAYY190 pKa = 10.02RR191 pKa = 11.84AMTPDD196 pKa = 3.43QEE198 pKa = 5.72SIDD201 pKa = 4.03ATKK204 pKa = 9.72MAYY207 pKa = 9.67NPKK210 pKa = 9.32SQARR214 pKa = 11.84RR215 pKa = 11.84DD216 pKa = 3.6LVSILRR222 pKa = 11.84KK223 pKa = 9.94NISFGGFKK231 pKa = 10.66SLEE234 pKa = 4.18DD235 pKa = 3.89FLNN238 pKa = 3.66
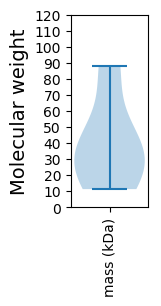
Molecular weight: 27.39 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q66096-2|RDRP-2_CRSVL Isoform of Q66096 Isoform protein p27 of RNA-directed RNA polymerase OS=Carnation ringspot virus (isolate Lommel) OX=652597 GN=ORF1 PE=4 SV=2
MM1 pKa = 7.43LSSYY5 pKa = 10.43LRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84EE10 pKa = 4.04PTRR13 pKa = 11.84HH14 pKa = 5.95KK15 pKa = 10.76SVIRR19 pKa = 11.84ILRR22 pKa = 11.84NRR24 pKa = 11.84QSATLVGGGRR34 pKa = 11.84RR35 pKa = 11.84VGFSLGVVPKK45 pKa = 10.85DD46 pKa = 4.46NLPTWWRR53 pKa = 11.84HH54 pKa = 3.83QACCGVIEE62 pKa = 4.69SRR64 pKa = 11.84TLVLAVVKK72 pKa = 9.47FVSVLFFCLYY82 pKa = 9.91VGSVCVTRR90 pKa = 11.84TDD92 pKa = 3.65GSRR95 pKa = 11.84KK96 pKa = 8.85PAVKK100 pKa = 10.3
MM1 pKa = 7.43LSSYY5 pKa = 10.43LRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84EE10 pKa = 4.04PTRR13 pKa = 11.84HH14 pKa = 5.95KK15 pKa = 10.76SVIRR19 pKa = 11.84ILRR22 pKa = 11.84NRR24 pKa = 11.84QSATLVGGGRR34 pKa = 11.84RR35 pKa = 11.84VGFSLGVVPKK45 pKa = 10.85DD46 pKa = 4.46NLPTWWRR53 pKa = 11.84HH54 pKa = 3.83QACCGVIEE62 pKa = 4.69SRR64 pKa = 11.84TLVLAVVKK72 pKa = 9.47FVSVLFFCLYY82 pKa = 9.91VGSVCVTRR90 pKa = 11.84TDD92 pKa = 3.65GSRR95 pKa = 11.84KK96 pKa = 8.85PAVKK100 pKa = 10.3
Molecular weight: 11.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1756 |
100 |
769 |
351.2 |
39.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.777 ± 0.552 | 2.335 ± 0.435 |
6.15 ± 0.52 | 4.841 ± 1.11 |
4.385 ± 0.524 | 5.41 ± 0.347 |
1.822 ± 0.264 | 5.068 ± 0.508 |
6.492 ± 0.708 | 7.802 ± 0.284 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.018 ± 0.425 | 4.1 ± 0.367 |
5.011 ± 0.535 | 3.189 ± 0.386 |
7.062 ± 0.533 | 8.599 ± 0.751 |
5.353 ± 1.388 | 7.973 ± 1.001 |
1.595 ± 0.19 | 3.018 ± 0.38 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
