
Bat coronavirus HKU4 (BtCoV) (BtCoV/HKU4/2004)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Betacoronavirus; Merbecovirus
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
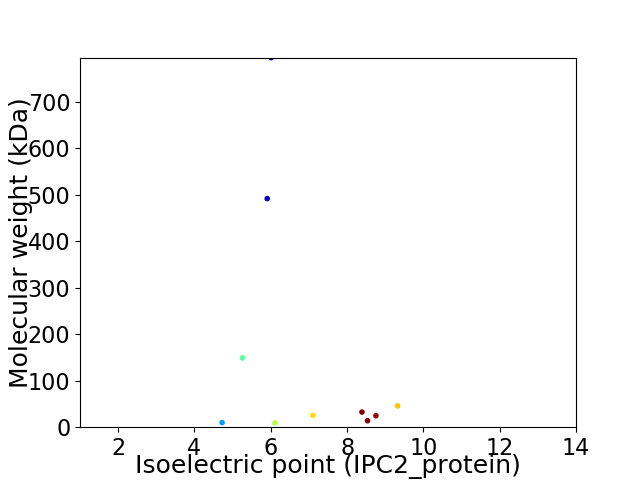
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
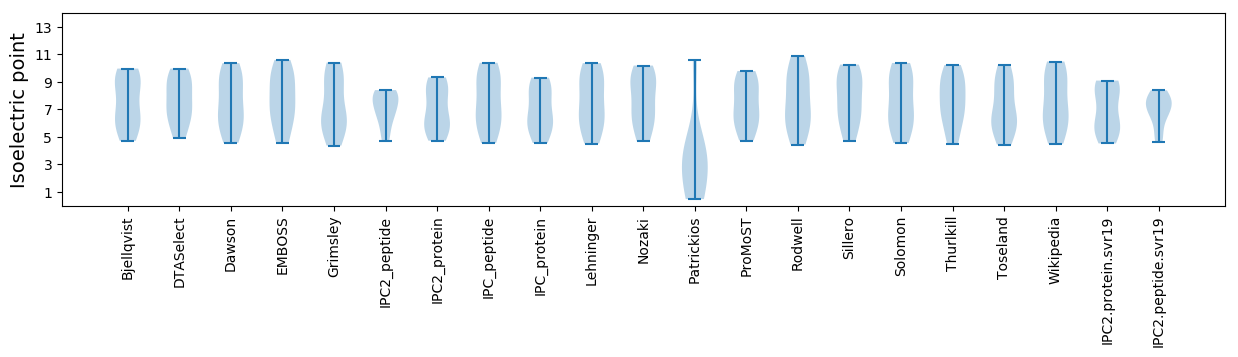
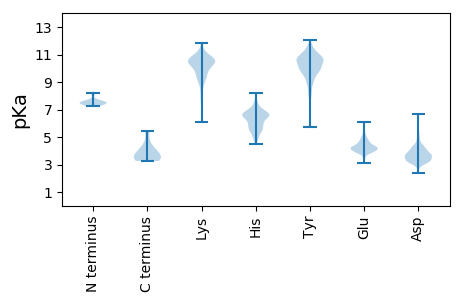
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|A3EX96|NS3B_BCHK4 Non-structural protein 3b OS=Bat coronavirus HKU4 OX=694007 GN=3b PE=3 SV=1
MM1 pKa = 7.34VSFNATAILLLLLANAFSKK20 pKa = 10.33PLYY23 pKa = 10.14VPEE26 pKa = 4.66HH27 pKa = 6.51CGGMSGTLFQACIRR41 pKa = 11.84QTMVDD46 pKa = 3.47TTGMYY51 pKa = 9.04TNSAMSHH58 pKa = 6.87DD59 pKa = 4.37GVTIPFDD66 pKa = 3.73RR67 pKa = 11.84DD68 pKa = 3.79GIVHH72 pKa = 6.91EE73 pKa = 4.39DD74 pKa = 3.99HH75 pKa = 6.3YY76 pKa = 10.19TEE78 pKa = 4.55TNPTPLFDD86 pKa = 5.08AGFSVV91 pKa = 4.1
MM1 pKa = 7.34VSFNATAILLLLLANAFSKK20 pKa = 10.33PLYY23 pKa = 10.14VPEE26 pKa = 4.66HH27 pKa = 6.51CGGMSGTLFQACIRR41 pKa = 11.84QTMVDD46 pKa = 3.47TTGMYY51 pKa = 9.04TNSAMSHH58 pKa = 6.87DD59 pKa = 4.37GVTIPFDD66 pKa = 3.73RR67 pKa = 11.84DD68 pKa = 3.79GIVHH72 pKa = 6.91EE73 pKa = 4.39DD74 pKa = 3.99HH75 pKa = 6.3YY76 pKa = 10.19TEE78 pKa = 4.55TNPTPLFDD86 pKa = 5.08AGFSVV91 pKa = 4.1
Molecular weight: 9.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P0C6T4|R1A_BCHK4 Replicase polyprotein 1a OS=Bat coronavirus HKU4 OX=694007 GN=1a PE=1 SV=1
MM1 pKa = 7.35ATPAAPRR8 pKa = 11.84TISFADD14 pKa = 3.62NNDD17 pKa = 3.4NQPNQQQRR25 pKa = 11.84GRR27 pKa = 11.84GRR29 pKa = 11.84NPKK32 pKa = 9.25PRR34 pKa = 11.84PAPNNTVSWYY44 pKa = 9.65TGLTQHH50 pKa = 6.52GKK52 pKa = 9.78NPLAFPPGQGVPLNANSTTAQNAGYY77 pKa = 9.31WRR79 pKa = 11.84RR80 pKa = 11.84QDD82 pKa = 3.96RR83 pKa = 11.84KK84 pKa = 9.42INTGNGVKK92 pKa = 10.0QLAPRR97 pKa = 11.84WFFYY101 pKa = 9.92YY102 pKa = 10.27TGTGPEE108 pKa = 3.84ANLPFRR114 pKa = 11.84SVKK117 pKa = 10.67DD118 pKa = 3.88GIVWVYY124 pKa = 11.15EE125 pKa = 4.21EE126 pKa = 5.03GATDD130 pKa = 3.58APSVFGTRR138 pKa = 11.84NPANDD143 pKa = 3.63AAIVCQFAPGTLIPKK158 pKa = 9.26NFHH161 pKa = 6.53IEE163 pKa = 4.18GTGGNSQSSSRR174 pKa = 11.84ASSNSRR180 pKa = 11.84NSSRR184 pKa = 11.84SSSRR188 pKa = 11.84GGRR191 pKa = 11.84STSNSRR197 pKa = 11.84GTSPVSHH204 pKa = 6.3GVGSAEE210 pKa = 4.25SLAALPLLLDD220 pKa = 3.75LQKK223 pKa = 11.08RR224 pKa = 11.84LADD227 pKa = 3.86LEE229 pKa = 4.54SGKK232 pKa = 10.61SKK234 pKa = 10.4QPKK237 pKa = 9.57VVTKK241 pKa = 10.49KK242 pKa = 10.56DD243 pKa = 2.83AAAAKK248 pKa = 10.32NKK250 pKa = 8.56MRR252 pKa = 11.84HH253 pKa = 5.39KK254 pKa = 10.51RR255 pKa = 11.84VATKK259 pKa = 10.64GFNVTQAFGLRR270 pKa = 11.84GPGPLQGNFGDD281 pKa = 3.68MNYY284 pKa = 10.59NKK286 pKa = 10.08FGTEE290 pKa = 3.93DD291 pKa = 3.63PRR293 pKa = 11.84WPQMAEE299 pKa = 3.91LAPSASAFMSMSQFKK314 pKa = 8.91LTHH317 pKa = 5.98QSNDD321 pKa = 3.46DD322 pKa = 3.85KK323 pKa = 11.68GDD325 pKa = 4.31PIYY328 pKa = 10.57FLSYY332 pKa = 10.6SGAIKK337 pKa = 10.63LDD339 pKa = 3.72PKK341 pKa = 10.55NPNYY345 pKa = 10.35KK346 pKa = 9.79KK347 pKa = 9.77WLEE350 pKa = 3.89LLEE353 pKa = 4.58SNIDD357 pKa = 3.4AYY359 pKa = 9.71KK360 pKa = 10.21TFPKK364 pKa = 10.18KK365 pKa = 9.25EE366 pKa = 4.1RR367 pKa = 11.84KK368 pKa = 9.31PKK370 pKa = 7.46TTEE373 pKa = 4.02DD374 pKa = 3.6GAVASSSASQMEE386 pKa = 4.42DD387 pKa = 2.37VDD389 pKa = 5.05AKK391 pKa = 10.18PQRR394 pKa = 11.84KK395 pKa = 8.32PKK397 pKa = 10.45SRR399 pKa = 11.84VAGSITMRR407 pKa = 11.84SGSLPALQDD416 pKa = 3.21VTFDD420 pKa = 3.85SEE422 pKa = 4.18AA423 pKa = 3.72
MM1 pKa = 7.35ATPAAPRR8 pKa = 11.84TISFADD14 pKa = 3.62NNDD17 pKa = 3.4NQPNQQQRR25 pKa = 11.84GRR27 pKa = 11.84GRR29 pKa = 11.84NPKK32 pKa = 9.25PRR34 pKa = 11.84PAPNNTVSWYY44 pKa = 9.65TGLTQHH50 pKa = 6.52GKK52 pKa = 9.78NPLAFPPGQGVPLNANSTTAQNAGYY77 pKa = 9.31WRR79 pKa = 11.84RR80 pKa = 11.84QDD82 pKa = 3.96RR83 pKa = 11.84KK84 pKa = 9.42INTGNGVKK92 pKa = 10.0QLAPRR97 pKa = 11.84WFFYY101 pKa = 9.92YY102 pKa = 10.27TGTGPEE108 pKa = 3.84ANLPFRR114 pKa = 11.84SVKK117 pKa = 10.67DD118 pKa = 3.88GIVWVYY124 pKa = 11.15EE125 pKa = 4.21EE126 pKa = 5.03GATDD130 pKa = 3.58APSVFGTRR138 pKa = 11.84NPANDD143 pKa = 3.63AAIVCQFAPGTLIPKK158 pKa = 9.26NFHH161 pKa = 6.53IEE163 pKa = 4.18GTGGNSQSSSRR174 pKa = 11.84ASSNSRR180 pKa = 11.84NSSRR184 pKa = 11.84SSSRR188 pKa = 11.84GGRR191 pKa = 11.84STSNSRR197 pKa = 11.84GTSPVSHH204 pKa = 6.3GVGSAEE210 pKa = 4.25SLAALPLLLDD220 pKa = 3.75LQKK223 pKa = 11.08RR224 pKa = 11.84LADD227 pKa = 3.86LEE229 pKa = 4.54SGKK232 pKa = 10.61SKK234 pKa = 10.4QPKK237 pKa = 9.57VVTKK241 pKa = 10.49KK242 pKa = 10.56DD243 pKa = 2.83AAAAKK248 pKa = 10.32NKK250 pKa = 8.56MRR252 pKa = 11.84HH253 pKa = 5.39KK254 pKa = 10.51RR255 pKa = 11.84VATKK259 pKa = 10.64GFNVTQAFGLRR270 pKa = 11.84GPGPLQGNFGDD281 pKa = 3.68MNYY284 pKa = 10.59NKK286 pKa = 10.08FGTEE290 pKa = 3.93DD291 pKa = 3.63PRR293 pKa = 11.84WPQMAEE299 pKa = 3.91LAPSASAFMSMSQFKK314 pKa = 8.91LTHH317 pKa = 5.98QSNDD321 pKa = 3.46DD322 pKa = 3.85KK323 pKa = 11.68GDD325 pKa = 4.31PIYY328 pKa = 10.57FLSYY332 pKa = 10.6SGAIKK337 pKa = 10.63LDD339 pKa = 3.72PKK341 pKa = 10.55NPNYY345 pKa = 10.35KK346 pKa = 9.79KK347 pKa = 9.77WLEE350 pKa = 3.89LLEE353 pKa = 4.58SNIDD357 pKa = 3.4AYY359 pKa = 9.71KK360 pKa = 10.21TFPKK364 pKa = 10.18KK365 pKa = 9.25EE366 pKa = 4.1RR367 pKa = 11.84KK368 pKa = 9.31PKK370 pKa = 7.46TTEE373 pKa = 4.02DD374 pKa = 3.6GAVASSSASQMEE386 pKa = 4.42DD387 pKa = 2.37VDD389 pKa = 5.05AKK391 pKa = 10.18PQRR394 pKa = 11.84KK395 pKa = 8.32PKK397 pKa = 10.45SRR399 pKa = 11.84VAGSITMRR407 pKa = 11.84SGSLPALQDD416 pKa = 3.21VTFDD420 pKa = 3.85SEE422 pKa = 4.18AA423 pKa = 3.72
Molecular weight: 45.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
14351 |
82 |
7119 |
1435.1 |
159.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.073 ± 0.259 | 3.052 ± 0.292 |
5.477 ± 0.319 | 3.846 ± 0.182 |
4.885 ± 0.222 | 5.937 ± 0.256 |
1.93 ± 0.146 | 4.94 ± 0.285 |
5.372 ± 0.366 | 9.553 ± 0.344 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.251 ± 0.127 | 5.526 ± 0.142 |
3.944 ± 0.617 | 3.087 ± 0.388 |
3.589 ± 0.29 | 7.449 ± 0.719 |
6.759 ± 0.266 | 9.198 ± 0.855 |
1.178 ± 0.095 | 4.954 ± 0.345 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
