
Halorubrum sp. CBA1125
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Halorubraceae; Halorubrum; unclassified Halorubrum
Average proteome isoelectric point is 4.89
Get precalculated fractions of proteins
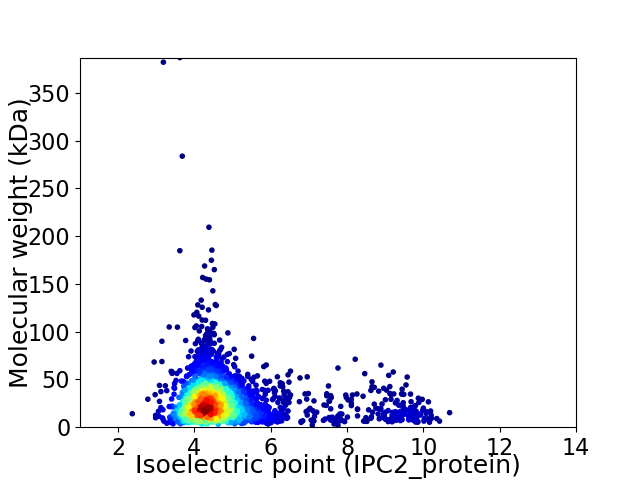
Virtual 2D-PAGE plot for 2859 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6A9T8R6|A0A6A9T8R6_9EURY Uncharacterized protein OS=Halorubrum sp. CBA1125 OX=2668072 GN=GJ633_08520 PE=4 SV=1
MM1 pKa = 7.6GLTDD5 pKa = 3.52TLAARR10 pKa = 11.84FDD12 pKa = 4.07VAEE15 pKa = 4.73HH16 pKa = 6.37GSDD19 pKa = 3.11VRR21 pKa = 11.84TEE23 pKa = 4.09VVAGITTFLAMSYY36 pKa = 9.92IIVVNPTILSNAIQIEE52 pKa = 5.14GYY54 pKa = 9.23SQGEE58 pKa = 4.73VIQMIAIATVLSSAVATLVMAFYY81 pKa = 11.22ANRR84 pKa = 11.84PFGLAPGLGLNAFFAFTVVLGLGIPWEE111 pKa = 4.16TALAAVFVEE120 pKa = 4.94GVLFMVLTAIGARR133 pKa = 11.84EE134 pKa = 3.97YY135 pKa = 10.26IIQLFPEE142 pKa = 4.3PVKK145 pKa = 10.61RR146 pKa = 11.84SVGAGIGLFLLFIGFQEE163 pKa = 4.06LQIIVNDD170 pKa = 3.38EE171 pKa = 4.05ATFVTLGQIFANPWAILGVLGLVLTFVLWARR202 pKa = 11.84DD203 pKa = 3.22ITGAIVIGILTTAAAGWALTLGGVFDD229 pKa = 5.35RR230 pKa = 11.84GTITPEE236 pKa = 3.79SLPAAQYY243 pKa = 10.51DD244 pKa = 3.53ISPLVGAFVEE254 pKa = 4.82GLADD258 pKa = 3.7VDD260 pKa = 4.21PLTFTLVVFTFFFVDD275 pKa = 4.8FFDD278 pKa = 3.96TAGTLIGVSQFGDD291 pKa = 3.54FLDD294 pKa = 5.43DD295 pKa = 4.54EE296 pKa = 5.5GDD298 pKa = 3.73LPEE301 pKa = 4.25MDD303 pKa = 4.4RR304 pKa = 11.84PLMADD309 pKa = 2.98AVGTTFGAIVGTSTVTTYY327 pKa = 10.34IEE329 pKa = 4.18SSTGVEE335 pKa = 3.77EE336 pKa = 4.63GGRR339 pKa = 11.84TGLTALVVAIAFLASLALIPVVAAIPAYY367 pKa = 10.36ASFIALIVVGVIMLQGLVEE386 pKa = 4.34VDD388 pKa = 2.88WTDD391 pKa = 3.42PAWAVSAGLTVTVMPLAYY409 pKa = 10.38SIADD413 pKa = 3.53GLAAGIVAYY422 pKa = 8.68PIIKK426 pKa = 9.77VAIGEE431 pKa = 4.24YY432 pKa = 10.53DD433 pKa = 3.63DD434 pKa = 4.58VAAGQYY440 pKa = 10.29VIAAVLAAYY449 pKa = 9.77YY450 pKa = 10.07VFQTSGLILL459 pKa = 3.89
MM1 pKa = 7.6GLTDD5 pKa = 3.52TLAARR10 pKa = 11.84FDD12 pKa = 4.07VAEE15 pKa = 4.73HH16 pKa = 6.37GSDD19 pKa = 3.11VRR21 pKa = 11.84TEE23 pKa = 4.09VVAGITTFLAMSYY36 pKa = 9.92IIVVNPTILSNAIQIEE52 pKa = 5.14GYY54 pKa = 9.23SQGEE58 pKa = 4.73VIQMIAIATVLSSAVATLVMAFYY81 pKa = 11.22ANRR84 pKa = 11.84PFGLAPGLGLNAFFAFTVVLGLGIPWEE111 pKa = 4.16TALAAVFVEE120 pKa = 4.94GVLFMVLTAIGARR133 pKa = 11.84EE134 pKa = 3.97YY135 pKa = 10.26IIQLFPEE142 pKa = 4.3PVKK145 pKa = 10.61RR146 pKa = 11.84SVGAGIGLFLLFIGFQEE163 pKa = 4.06LQIIVNDD170 pKa = 3.38EE171 pKa = 4.05ATFVTLGQIFANPWAILGVLGLVLTFVLWARR202 pKa = 11.84DD203 pKa = 3.22ITGAIVIGILTTAAAGWALTLGGVFDD229 pKa = 5.35RR230 pKa = 11.84GTITPEE236 pKa = 3.79SLPAAQYY243 pKa = 10.51DD244 pKa = 3.53ISPLVGAFVEE254 pKa = 4.82GLADD258 pKa = 3.7VDD260 pKa = 4.21PLTFTLVVFTFFFVDD275 pKa = 4.8FFDD278 pKa = 3.96TAGTLIGVSQFGDD291 pKa = 3.54FLDD294 pKa = 5.43DD295 pKa = 4.54EE296 pKa = 5.5GDD298 pKa = 3.73LPEE301 pKa = 4.25MDD303 pKa = 4.4RR304 pKa = 11.84PLMADD309 pKa = 2.98AVGTTFGAIVGTSTVTTYY327 pKa = 10.34IEE329 pKa = 4.18SSTGVEE335 pKa = 3.77EE336 pKa = 4.63GGRR339 pKa = 11.84TGLTALVVAIAFLASLALIPVVAAIPAYY367 pKa = 10.36ASFIALIVVGVIMLQGLVEE386 pKa = 4.34VDD388 pKa = 2.88WTDD391 pKa = 3.42PAWAVSAGLTVTVMPLAYY409 pKa = 10.38SIADD413 pKa = 3.53GLAAGIVAYY422 pKa = 8.68PIIKK426 pKa = 9.77VAIGEE431 pKa = 4.24YY432 pKa = 10.53DD433 pKa = 3.63DD434 pKa = 4.58VAAGQYY440 pKa = 10.29VIAAVLAAYY449 pKa = 9.77YY450 pKa = 10.07VFQTSGLILL459 pKa = 3.89
Molecular weight: 48.03 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6A9T5C4|A0A6A9T5C4_9EURY Phosphoesterase OS=Halorubrum sp. CBA1125 OX=2668072 GN=GJ633_06600 PE=3 SV=1
MM1 pKa = 7.22TVRR4 pKa = 11.84KK5 pKa = 9.12AAFSPAVPSAVVAVALAAGVVVSLDD30 pKa = 3.11ARR32 pKa = 11.84EE33 pKa = 4.18AVLATLAVQGGGLAVVGIGVASRR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84GHH60 pKa = 6.99RR61 pKa = 11.84ILGTALAVVGAGVIVGAFDD80 pKa = 4.52LFVDD84 pKa = 3.89EE85 pKa = 5.12RR86 pKa = 11.84HH87 pKa = 5.87ALSYY91 pKa = 9.26TIRR94 pKa = 11.84FLPGLVGIPLLAGTLLPVWASGSRR118 pKa = 11.84LVCKK122 pKa = 10.43LGTGGVFLTVVFSGLFSAVGEE143 pKa = 4.05LHH145 pKa = 6.88LLLAAVATVVAWDD158 pKa = 3.61AADD161 pKa = 3.48NAVGIGEE168 pKa = 4.3HH169 pKa = 6.75LGRR172 pKa = 11.84AATSWRR178 pKa = 11.84LQVTHH183 pKa = 5.7TTGSAAVGLAGVAGIHH199 pKa = 6.01LVRR202 pKa = 11.84HH203 pKa = 5.33VAVSDD208 pKa = 3.94LSLAAFALLFVALLLLLAAIRR229 pKa = 11.84GG230 pKa = 3.82
MM1 pKa = 7.22TVRR4 pKa = 11.84KK5 pKa = 9.12AAFSPAVPSAVVAVALAAGVVVSLDD30 pKa = 3.11ARR32 pKa = 11.84EE33 pKa = 4.18AVLATLAVQGGGLAVVGIGVASRR56 pKa = 11.84RR57 pKa = 11.84RR58 pKa = 11.84GHH60 pKa = 6.99RR61 pKa = 11.84ILGTALAVVGAGVIVGAFDD80 pKa = 4.52LFVDD84 pKa = 3.89EE85 pKa = 5.12RR86 pKa = 11.84HH87 pKa = 5.87ALSYY91 pKa = 9.26TIRR94 pKa = 11.84FLPGLVGIPLLAGTLLPVWASGSRR118 pKa = 11.84LVCKK122 pKa = 10.43LGTGGVFLTVVFSGLFSAVGEE143 pKa = 4.05LHH145 pKa = 6.88LLLAAVATVVAWDD158 pKa = 3.61AADD161 pKa = 3.48NAVGIGEE168 pKa = 4.3HH169 pKa = 6.75LGRR172 pKa = 11.84AATSWRR178 pKa = 11.84LQVTHH183 pKa = 5.7TTGSAAVGLAGVAGIHH199 pKa = 6.01LVRR202 pKa = 11.84HH203 pKa = 5.33VAVSDD208 pKa = 3.94LSLAAFALLFVALLLLLAAIRR229 pKa = 11.84GG230 pKa = 3.82
Molecular weight: 23.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
754458 |
15 |
3720 |
263.9 |
28.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.074 ± 0.074 | 0.71 ± 0.016 |
8.769 ± 0.063 | 8.419 ± 0.064 |
3.283 ± 0.03 | 8.534 ± 0.058 |
1.949 ± 0.026 | 4.181 ± 0.035 |
1.787 ± 0.032 | 8.613 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.658 ± 0.025 | 2.372 ± 0.04 |
4.729 ± 0.034 | 2.354 ± 0.028 |
6.716 ± 0.06 | 5.395 ± 0.048 |
6.445 ± 0.069 | 9.216 ± 0.055 |
1.115 ± 0.018 | 2.681 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
