
McMurdo Ice Shelf pond-associated circular DNA virus-5
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.45
Get precalculated fractions of proteins
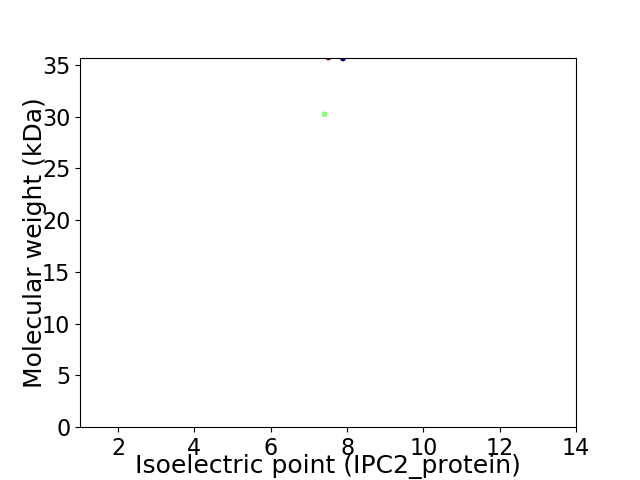
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075M3Q1|A0A075M3Q1_9VIRU Putative capsid protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-5 OX=1521389 PE=4 SV=1
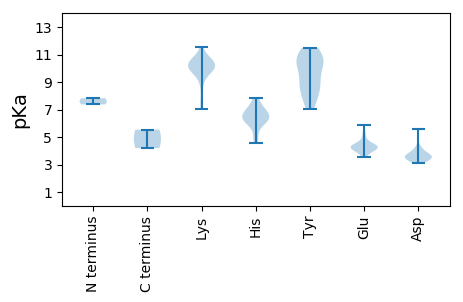
MM1 pKa = 7.82SDD3 pKa = 3.4FSEE6 pKa = 4.38YY7 pKa = 9.87TQISDD12 pKa = 3.45LSTYY16 pKa = 7.52EE17 pKa = 3.86WPSGSRR23 pKa = 11.84RR24 pKa = 11.84PPTEE28 pKa = 3.53NARR31 pKa = 11.84TTLDD35 pKa = 3.18EE36 pKa = 5.45LMNEE40 pKa = 4.44LLNIEE45 pKa = 4.28EE46 pKa = 5.06PEE48 pKa = 4.23LSHH51 pKa = 6.37EE52 pKa = 4.4TALLMNVYY60 pKa = 8.24TQEE63 pKa = 4.04TLLRR67 pKa = 11.84DD68 pKa = 3.68YY69 pKa = 11.45VNRR72 pKa = 11.84STWLAPEE79 pKa = 4.33DD80 pKa = 4.06LKK82 pKa = 11.52NALTSWYY89 pKa = 10.0YY90 pKa = 10.75HH91 pKa = 6.0MNIIWYY97 pKa = 8.51PSKK100 pKa = 10.04TPLTSTLSFLRR111 pKa = 11.84RR112 pKa = 11.84YY113 pKa = 7.64RR114 pKa = 11.84TSKK117 pKa = 10.57LGNNAQIRR125 pKa = 11.84YY126 pKa = 8.75LLLKK130 pKa = 10.04EE131 pKa = 4.03SHH133 pKa = 6.29LTLEE137 pKa = 4.35ARR139 pKa = 11.84EE140 pKa = 4.06QFYY143 pKa = 9.12MICQPEE149 pKa = 4.32THH151 pKa = 6.76ISHH154 pKa = 6.36YY155 pKa = 10.32PSTIQQNQASIPKK168 pKa = 9.74DD169 pKa = 3.43VQDD172 pKa = 4.26LVRR175 pKa = 11.84PVLKK179 pKa = 10.5SRR181 pKa = 11.84KK182 pKa = 7.03QTLNKK187 pKa = 10.14CPLTSLPISLQVRR200 pKa = 11.84AVGLFAIEE208 pKa = 4.67RR209 pKa = 11.84LTLPLASSLVVVTKK223 pKa = 10.33CSATLPTIPNCSQLFQKK240 pKa = 10.34QSCQNCQCCKK250 pKa = 8.07TTLSYY255 pKa = 11.04TRR257 pKa = 11.84TITYY261 pKa = 9.34HH262 pKa = 5.54
MM1 pKa = 7.82SDD3 pKa = 3.4FSEE6 pKa = 4.38YY7 pKa = 9.87TQISDD12 pKa = 3.45LSTYY16 pKa = 7.52EE17 pKa = 3.86WPSGSRR23 pKa = 11.84RR24 pKa = 11.84PPTEE28 pKa = 3.53NARR31 pKa = 11.84TTLDD35 pKa = 3.18EE36 pKa = 5.45LMNEE40 pKa = 4.44LLNIEE45 pKa = 4.28EE46 pKa = 5.06PEE48 pKa = 4.23LSHH51 pKa = 6.37EE52 pKa = 4.4TALLMNVYY60 pKa = 8.24TQEE63 pKa = 4.04TLLRR67 pKa = 11.84DD68 pKa = 3.68YY69 pKa = 11.45VNRR72 pKa = 11.84STWLAPEE79 pKa = 4.33DD80 pKa = 4.06LKK82 pKa = 11.52NALTSWYY89 pKa = 10.0YY90 pKa = 10.75HH91 pKa = 6.0MNIIWYY97 pKa = 8.51PSKK100 pKa = 10.04TPLTSTLSFLRR111 pKa = 11.84RR112 pKa = 11.84YY113 pKa = 7.64RR114 pKa = 11.84TSKK117 pKa = 10.57LGNNAQIRR125 pKa = 11.84YY126 pKa = 8.75LLLKK130 pKa = 10.04EE131 pKa = 4.03SHH133 pKa = 6.29LTLEE137 pKa = 4.35ARR139 pKa = 11.84EE140 pKa = 4.06QFYY143 pKa = 9.12MICQPEE149 pKa = 4.32THH151 pKa = 6.76ISHH154 pKa = 6.36YY155 pKa = 10.32PSTIQQNQASIPKK168 pKa = 9.74DD169 pKa = 3.43VQDD172 pKa = 4.26LVRR175 pKa = 11.84PVLKK179 pKa = 10.5SRR181 pKa = 11.84KK182 pKa = 7.03QTLNKK187 pKa = 10.14CPLTSLPISLQVRR200 pKa = 11.84AVGLFAIEE208 pKa = 4.67RR209 pKa = 11.84LTLPLASSLVVVTKK223 pKa = 10.33CSATLPTIPNCSQLFQKK240 pKa = 10.34QSCQNCQCCKK250 pKa = 8.07TTLSYY255 pKa = 11.04TRR257 pKa = 11.84TITYY261 pKa = 9.34HH262 pKa = 5.54
Molecular weight: 30.22 kDa
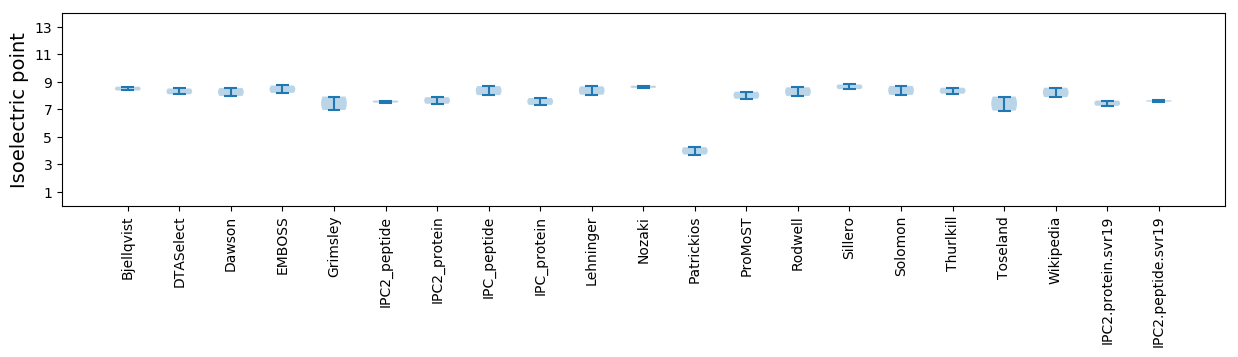
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075M3Q1|A0A075M3Q1_9VIRU Putative capsid protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-5 OX=1521389 PE=4 SV=1
MM1 pKa = 7.37SRR3 pKa = 11.84AKK5 pKa = 9.88HH6 pKa = 4.57WCFTVNNYY14 pKa = 8.25TDD16 pKa = 3.76EE17 pKa = 5.91DD18 pKa = 3.51IHH20 pKa = 7.87KK21 pKa = 10.23LSKK24 pKa = 10.81ASLLLQPLVSSCIYY38 pKa = 8.45QQEE41 pKa = 4.55VPGQEE46 pKa = 4.23SATPGTPHH54 pKa = 6.33LQGFISFKK62 pKa = 9.72TKK64 pKa = 10.47QSFKK68 pKa = 9.21FTKK71 pKa = 10.52NLVSDD76 pKa = 4.24RR77 pKa = 11.84AHH79 pKa = 6.78VEE81 pKa = 4.16VAKK84 pKa = 8.93GTPQQNRR91 pKa = 11.84IYY93 pKa = 10.38CSKK96 pKa = 11.11AKK98 pKa = 9.91DD99 pKa = 3.47RR100 pKa = 11.84KK101 pKa = 10.01IGTEE105 pKa = 3.7VFTYY109 pKa = 10.91GEE111 pKa = 4.17QPKK114 pKa = 10.28LLPGKK119 pKa = 10.59RR120 pKa = 11.84NDD122 pKa = 3.72LYY124 pKa = 11.48AFQQYY129 pKa = 9.21VKK131 pKa = 9.89EE132 pKa = 4.4GNIQTRR138 pKa = 11.84DD139 pKa = 3.1ILEE142 pKa = 4.01NHH144 pKa = 6.71ASVAARR150 pKa = 11.84YY151 pKa = 7.59PRR153 pKa = 11.84YY154 pKa = 8.67VRR156 pKa = 11.84EE157 pKa = 4.18YY158 pKa = 9.22IDD160 pKa = 5.59LYY162 pKa = 10.96VNPPEE167 pKa = 5.04VPDD170 pKa = 3.71HH171 pKa = 7.11PLYY174 pKa = 10.69LWQEE178 pKa = 4.26TLTKK182 pKa = 10.08IITGPSDD189 pKa = 3.2DD190 pKa = 3.7RR191 pKa = 11.84QIIFVVDD198 pKa = 3.47EE199 pKa = 4.42VGNQGKK205 pKa = 7.32TWFAKK210 pKa = 9.89KK211 pKa = 9.87YY212 pKa = 10.1CRR214 pKa = 11.84AHH216 pKa = 6.85EE217 pKa = 4.39DD218 pKa = 3.49AQFLEE223 pKa = 4.64PGKK226 pKa = 10.4KK227 pKa = 9.85ADD229 pKa = 3.33MAYY232 pKa = 10.74ALNTDD237 pKa = 3.52LRR239 pKa = 11.84VLFLNVTRR247 pKa = 11.84QQVEE251 pKa = 3.82HH252 pKa = 6.17LQYY255 pKa = 11.43SFLEE259 pKa = 4.15AVKK262 pKa = 10.56DD263 pKa = 3.99GSVWSPKK270 pKa = 9.36YY271 pKa = 10.17EE272 pKa = 4.01SRR274 pKa = 11.84TKK276 pKa = 10.57HH277 pKa = 5.12LTQIPHH283 pKa = 6.13IVVMMNQDD291 pKa = 3.58PDD293 pKa = 3.77FQLLSKK299 pKa = 10.69DD300 pKa = 3.63RR301 pKa = 11.84YY302 pKa = 7.02HH303 pKa = 7.33TIYY306 pKa = 10.38II307 pKa = 4.19
MM1 pKa = 7.37SRR3 pKa = 11.84AKK5 pKa = 9.88HH6 pKa = 4.57WCFTVNNYY14 pKa = 8.25TDD16 pKa = 3.76EE17 pKa = 5.91DD18 pKa = 3.51IHH20 pKa = 7.87KK21 pKa = 10.23LSKK24 pKa = 10.81ASLLLQPLVSSCIYY38 pKa = 8.45QQEE41 pKa = 4.55VPGQEE46 pKa = 4.23SATPGTPHH54 pKa = 6.33LQGFISFKK62 pKa = 9.72TKK64 pKa = 10.47QSFKK68 pKa = 9.21FTKK71 pKa = 10.52NLVSDD76 pKa = 4.24RR77 pKa = 11.84AHH79 pKa = 6.78VEE81 pKa = 4.16VAKK84 pKa = 8.93GTPQQNRR91 pKa = 11.84IYY93 pKa = 10.38CSKK96 pKa = 11.11AKK98 pKa = 9.91DD99 pKa = 3.47RR100 pKa = 11.84KK101 pKa = 10.01IGTEE105 pKa = 3.7VFTYY109 pKa = 10.91GEE111 pKa = 4.17QPKK114 pKa = 10.28LLPGKK119 pKa = 10.59RR120 pKa = 11.84NDD122 pKa = 3.72LYY124 pKa = 11.48AFQQYY129 pKa = 9.21VKK131 pKa = 9.89EE132 pKa = 4.4GNIQTRR138 pKa = 11.84DD139 pKa = 3.1ILEE142 pKa = 4.01NHH144 pKa = 6.71ASVAARR150 pKa = 11.84YY151 pKa = 7.59PRR153 pKa = 11.84YY154 pKa = 8.67VRR156 pKa = 11.84EE157 pKa = 4.18YY158 pKa = 9.22IDD160 pKa = 5.59LYY162 pKa = 10.96VNPPEE167 pKa = 5.04VPDD170 pKa = 3.71HH171 pKa = 7.11PLYY174 pKa = 10.69LWQEE178 pKa = 4.26TLTKK182 pKa = 10.08IITGPSDD189 pKa = 3.2DD190 pKa = 3.7RR191 pKa = 11.84QIIFVVDD198 pKa = 3.47EE199 pKa = 4.42VGNQGKK205 pKa = 7.32TWFAKK210 pKa = 9.89KK211 pKa = 9.87YY212 pKa = 10.1CRR214 pKa = 11.84AHH216 pKa = 6.85EE217 pKa = 4.39DD218 pKa = 3.49AQFLEE223 pKa = 4.64PGKK226 pKa = 10.4KK227 pKa = 9.85ADD229 pKa = 3.33MAYY232 pKa = 10.74ALNTDD237 pKa = 3.52LRR239 pKa = 11.84VLFLNVTRR247 pKa = 11.84QQVEE251 pKa = 3.82HH252 pKa = 6.17LQYY255 pKa = 11.43SFLEE259 pKa = 4.15AVKK262 pKa = 10.56DD263 pKa = 3.99GSVWSPKK270 pKa = 9.36YY271 pKa = 10.17EE272 pKa = 4.01SRR274 pKa = 11.84TKK276 pKa = 10.57HH277 pKa = 5.12LTQIPHH283 pKa = 6.13IVVMMNQDD291 pKa = 3.58PDD293 pKa = 3.77FQLLSKK299 pKa = 10.69DD300 pKa = 3.63RR301 pKa = 11.84YY302 pKa = 7.02HH303 pKa = 7.33TIYY306 pKa = 10.38II307 pKa = 4.19
Molecular weight: 35.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
569 |
262 |
307 |
284.5 |
32.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.921 ± 0.465 | 2.109 ± 0.608 |
4.394 ± 1.109 | 5.8 ± 0.198 |
3.163 ± 0.808 | 2.812 ± 1.073 |
2.988 ± 0.449 | 5.097 ± 0.087 |
6.151 ± 1.257 | 10.721 ± 1.945 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.582 ± 0.21 | 4.394 ± 0.366 |
5.8 ± 0.198 | 6.678 ± 0.368 |
5.272 ± 0.292 | 7.557 ± 1.524 |
8.26 ± 1.563 | 5.624 ± 1.164 |
1.406 ± 0.078 | 5.272 ± 0.2 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
