
Stenomitos frigidus ULC18
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Leptolyngbyaceae; Stenomitos; Stenomitos frigidus
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
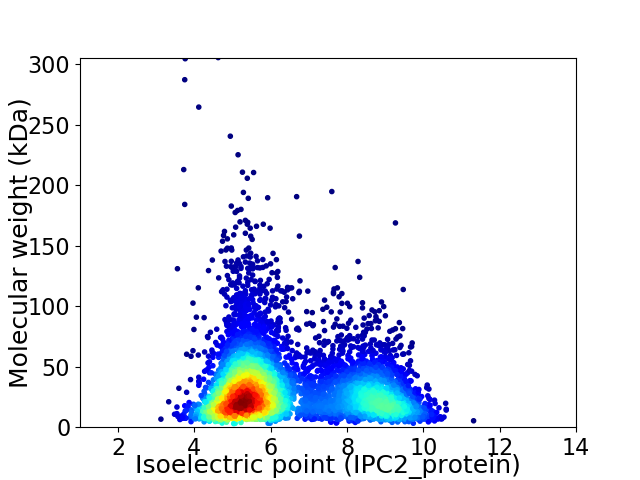
Virtual 2D-PAGE plot for 5916 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T1ED57|A0A2T1ED57_9CYAN Radical SAM protein OS=Stenomitos frigidus ULC18 OX=2107698 GN=C7B82_08340 PE=4 SV=1
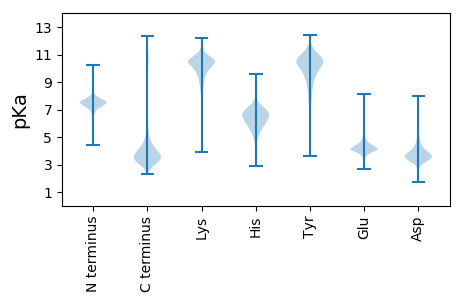
MM1 pKa = 7.51NEE3 pKa = 4.27HH4 pKa = 6.75IALPLCKK11 pKa = 9.84GLSMPVKK18 pKa = 10.24NFRR21 pKa = 11.84HH22 pKa = 4.68VRR24 pKa = 11.84KK25 pKa = 8.57ATVVKK30 pKa = 9.71TPDD33 pKa = 2.99GGQIKK38 pKa = 10.08LGSYY42 pKa = 10.34RR43 pKa = 11.84PDD45 pKa = 3.08KK46 pKa = 10.97KK47 pKa = 10.94NPNDD51 pKa = 3.42KK52 pKa = 9.64TYY54 pKa = 10.84KK55 pKa = 9.92SSRR58 pKa = 11.84STDD61 pKa = 3.61DD62 pKa = 4.18LPSKK66 pKa = 10.93VDD68 pKa = 3.41LRR70 pKa = 11.84PYY72 pKa = 6.64MTPVEE77 pKa = 4.14NQGNSNSCTANAMAGAYY94 pKa = 8.02EE95 pKa = 4.09YY96 pKa = 10.69LAKK99 pKa = 10.13RR100 pKa = 11.84VKK102 pKa = 10.71GSAHH106 pKa = 5.94DD107 pKa = 3.64VSRR110 pKa = 11.84LFIYY114 pKa = 10.89YY115 pKa = 9.9NARR118 pKa = 11.84EE119 pKa = 4.16LDD121 pKa = 3.47GATDD125 pKa = 3.79SDD127 pKa = 3.53EE128 pKa = 4.09GTYY131 pKa = 10.55LRR133 pKa = 11.84SCVKK137 pKa = 10.19VLKK140 pKa = 10.57KK141 pKa = 10.86YY142 pKa = 7.35GTCAEE147 pKa = 4.46TTWPFDD153 pKa = 2.94LHH155 pKa = 7.64RR156 pKa = 11.84IFEE159 pKa = 4.66PPHH162 pKa = 5.49EE163 pKa = 4.38SAYY166 pKa = 10.0TEE168 pKa = 3.9ATSFLIEE175 pKa = 3.62DD176 pKa = 4.48AYY178 pKa = 10.63RR179 pKa = 11.84VKK181 pKa = 10.88VDD183 pKa = 4.19LDD185 pKa = 3.47AMRR188 pKa = 11.84QCLADD193 pKa = 4.63GYY195 pKa = 9.62PFTFGLDD202 pKa = 3.46LFSSFQKK209 pKa = 10.63AGSKK213 pKa = 10.7GLVPMPDD220 pKa = 3.73PDD222 pKa = 4.73SEE224 pKa = 4.31QHH226 pKa = 6.87DD227 pKa = 4.14GGHH230 pKa = 7.12AMLCVGYY237 pKa = 10.66SDD239 pKa = 3.9ADD241 pKa = 3.36KK242 pKa = 11.46VFIVRR247 pKa = 11.84NSWGEE252 pKa = 3.51GWGDD256 pKa = 3.24HH257 pKa = 6.45GYY259 pKa = 11.03CYY261 pKa = 10.18IPYY264 pKa = 10.19DD265 pKa = 4.07YY266 pKa = 7.67MTNPDD271 pKa = 3.96LNGDD275 pKa = 3.87LWAIRR280 pKa = 11.84HH281 pKa = 5.35VADD284 pKa = 4.6ADD286 pKa = 3.45IDD288 pKa = 4.09LSQDD292 pKa = 2.87IQGEE296 pKa = 4.37RR297 pKa = 11.84TSLFSGATAAIAHH310 pKa = 6.68AMQLPRR316 pKa = 11.84ASRR319 pKa = 11.84TTEE322 pKa = 3.44YY323 pKa = 11.35DD324 pKa = 2.66NDD326 pKa = 3.82YY327 pKa = 8.97TVEE330 pKa = 3.99YY331 pKa = 9.63AAQSVFVNGGFIAISDD347 pKa = 4.11YY348 pKa = 11.62EE349 pKa = 4.59DD350 pKa = 4.75LDD352 pKa = 3.65SLYY355 pKa = 10.43FAEE358 pKa = 5.51YY359 pKa = 9.16EE360 pKa = 4.04DD361 pKa = 4.62EE362 pKa = 4.5YY363 pKa = 11.5EE364 pKa = 4.28EE365 pKa = 4.23FTLTEE370 pKa = 3.99TSEE373 pKa = 4.15EE374 pKa = 4.2TEE376 pKa = 3.93YY377 pKa = 11.03SYY379 pKa = 11.86EE380 pKa = 4.08EE381 pKa = 4.15TSEE384 pKa = 4.12EE385 pKa = 4.07TSEE388 pKa = 4.03EE389 pKa = 4.1TEE391 pKa = 3.91YY392 pKa = 11.03SYY394 pKa = 11.84EE395 pKa = 4.09EE396 pKa = 3.85TSEE399 pKa = 3.94EE400 pKa = 4.23SEE402 pKa = 4.07EE403 pKa = 4.35SEE405 pKa = 4.24EE406 pKa = 4.41TEE408 pKa = 4.07DD409 pKa = 3.95SYY411 pKa = 12.0EE412 pKa = 4.09EE413 pKa = 3.93EE414 pKa = 4.44SEE416 pKa = 4.24EE417 pKa = 4.61EE418 pKa = 3.94EE419 pKa = 5.51SEE421 pKa = 5.12DD422 pKa = 3.85EE423 pKa = 4.55DD424 pKa = 5.27EE425 pKa = 4.6EE426 pKa = 5.59SEE428 pKa = 5.23DD429 pKa = 3.71EE430 pKa = 4.19NEE432 pKa = 4.08EE433 pKa = 3.96DD434 pKa = 4.22SEE436 pKa = 4.53EE437 pKa = 4.55IEE439 pKa = 4.88DD440 pKa = 4.14SYY442 pKa = 12.12EE443 pKa = 3.87EE444 pKa = 4.71DD445 pKa = 3.94SEE447 pKa = 5.05DD448 pKa = 4.76DD449 pKa = 3.59EE450 pKa = 5.93SEE452 pKa = 4.41EE453 pKa = 4.04EE454 pKa = 4.17DD455 pKa = 4.25SEE457 pKa = 5.0EE458 pKa = 4.01EE459 pKa = 4.24DD460 pKa = 3.81SEE462 pKa = 4.15EE463 pKa = 4.42DD464 pKa = 3.67EE465 pKa = 4.42EE466 pKa = 5.6SEE468 pKa = 4.71EE469 pKa = 4.11EE470 pKa = 4.28DD471 pKa = 4.15SEE473 pKa = 4.34EE474 pKa = 4.33TEE476 pKa = 4.49DD477 pKa = 4.79SYY479 pKa = 12.2EE480 pKa = 4.03EE481 pKa = 4.51DD482 pKa = 3.6SEE484 pKa = 4.27EE485 pKa = 4.74DD486 pKa = 3.34EE487 pKa = 5.18SEE489 pKa = 4.39EE490 pKa = 3.93EE491 pKa = 4.07DD492 pKa = 3.86SEE494 pKa = 4.61EE495 pKa = 4.28EE496 pKa = 3.92EE497 pKa = 4.8SEE499 pKa = 4.25AEE501 pKa = 3.96PEE503 pKa = 4.14EE504 pKa = 4.39EE505 pKa = 4.61SEE507 pKa = 4.06EE508 pKa = 4.39DD509 pKa = 3.55YY510 pKa = 11.88GDD512 pKa = 4.47AEE514 pKa = 4.01ASYY517 pKa = 11.48DD518 pKa = 3.94EE519 pKa = 4.89GGEE522 pKa = 4.13DD523 pKa = 3.25EE524 pKa = 5.32GGGEE528 pKa = 4.39GEE530 pKa = 4.44EE531 pKa = 4.37EE532 pKa = 4.02
MM1 pKa = 7.51NEE3 pKa = 4.27HH4 pKa = 6.75IALPLCKK11 pKa = 9.84GLSMPVKK18 pKa = 10.24NFRR21 pKa = 11.84HH22 pKa = 4.68VRR24 pKa = 11.84KK25 pKa = 8.57ATVVKK30 pKa = 9.71TPDD33 pKa = 2.99GGQIKK38 pKa = 10.08LGSYY42 pKa = 10.34RR43 pKa = 11.84PDD45 pKa = 3.08KK46 pKa = 10.97KK47 pKa = 10.94NPNDD51 pKa = 3.42KK52 pKa = 9.64TYY54 pKa = 10.84KK55 pKa = 9.92SSRR58 pKa = 11.84STDD61 pKa = 3.61DD62 pKa = 4.18LPSKK66 pKa = 10.93VDD68 pKa = 3.41LRR70 pKa = 11.84PYY72 pKa = 6.64MTPVEE77 pKa = 4.14NQGNSNSCTANAMAGAYY94 pKa = 8.02EE95 pKa = 4.09YY96 pKa = 10.69LAKK99 pKa = 10.13RR100 pKa = 11.84VKK102 pKa = 10.71GSAHH106 pKa = 5.94DD107 pKa = 3.64VSRR110 pKa = 11.84LFIYY114 pKa = 10.89YY115 pKa = 9.9NARR118 pKa = 11.84EE119 pKa = 4.16LDD121 pKa = 3.47GATDD125 pKa = 3.79SDD127 pKa = 3.53EE128 pKa = 4.09GTYY131 pKa = 10.55LRR133 pKa = 11.84SCVKK137 pKa = 10.19VLKK140 pKa = 10.57KK141 pKa = 10.86YY142 pKa = 7.35GTCAEE147 pKa = 4.46TTWPFDD153 pKa = 2.94LHH155 pKa = 7.64RR156 pKa = 11.84IFEE159 pKa = 4.66PPHH162 pKa = 5.49EE163 pKa = 4.38SAYY166 pKa = 10.0TEE168 pKa = 3.9ATSFLIEE175 pKa = 3.62DD176 pKa = 4.48AYY178 pKa = 10.63RR179 pKa = 11.84VKK181 pKa = 10.88VDD183 pKa = 4.19LDD185 pKa = 3.47AMRR188 pKa = 11.84QCLADD193 pKa = 4.63GYY195 pKa = 9.62PFTFGLDD202 pKa = 3.46LFSSFQKK209 pKa = 10.63AGSKK213 pKa = 10.7GLVPMPDD220 pKa = 3.73PDD222 pKa = 4.73SEE224 pKa = 4.31QHH226 pKa = 6.87DD227 pKa = 4.14GGHH230 pKa = 7.12AMLCVGYY237 pKa = 10.66SDD239 pKa = 3.9ADD241 pKa = 3.36KK242 pKa = 11.46VFIVRR247 pKa = 11.84NSWGEE252 pKa = 3.51GWGDD256 pKa = 3.24HH257 pKa = 6.45GYY259 pKa = 11.03CYY261 pKa = 10.18IPYY264 pKa = 10.19DD265 pKa = 4.07YY266 pKa = 7.67MTNPDD271 pKa = 3.96LNGDD275 pKa = 3.87LWAIRR280 pKa = 11.84HH281 pKa = 5.35VADD284 pKa = 4.6ADD286 pKa = 3.45IDD288 pKa = 4.09LSQDD292 pKa = 2.87IQGEE296 pKa = 4.37RR297 pKa = 11.84TSLFSGATAAIAHH310 pKa = 6.68AMQLPRR316 pKa = 11.84ASRR319 pKa = 11.84TTEE322 pKa = 3.44YY323 pKa = 11.35DD324 pKa = 2.66NDD326 pKa = 3.82YY327 pKa = 8.97TVEE330 pKa = 3.99YY331 pKa = 9.63AAQSVFVNGGFIAISDD347 pKa = 4.11YY348 pKa = 11.62EE349 pKa = 4.59DD350 pKa = 4.75LDD352 pKa = 3.65SLYY355 pKa = 10.43FAEE358 pKa = 5.51YY359 pKa = 9.16EE360 pKa = 4.04DD361 pKa = 4.62EE362 pKa = 4.5YY363 pKa = 11.5EE364 pKa = 4.28EE365 pKa = 4.23FTLTEE370 pKa = 3.99TSEE373 pKa = 4.15EE374 pKa = 4.2TEE376 pKa = 3.93YY377 pKa = 11.03SYY379 pKa = 11.86EE380 pKa = 4.08EE381 pKa = 4.15TSEE384 pKa = 4.12EE385 pKa = 4.07TSEE388 pKa = 4.03EE389 pKa = 4.1TEE391 pKa = 3.91YY392 pKa = 11.03SYY394 pKa = 11.84EE395 pKa = 4.09EE396 pKa = 3.85TSEE399 pKa = 3.94EE400 pKa = 4.23SEE402 pKa = 4.07EE403 pKa = 4.35SEE405 pKa = 4.24EE406 pKa = 4.41TEE408 pKa = 4.07DD409 pKa = 3.95SYY411 pKa = 12.0EE412 pKa = 4.09EE413 pKa = 3.93EE414 pKa = 4.44SEE416 pKa = 4.24EE417 pKa = 4.61EE418 pKa = 3.94EE419 pKa = 5.51SEE421 pKa = 5.12DD422 pKa = 3.85EE423 pKa = 4.55DD424 pKa = 5.27EE425 pKa = 4.6EE426 pKa = 5.59SEE428 pKa = 5.23DD429 pKa = 3.71EE430 pKa = 4.19NEE432 pKa = 4.08EE433 pKa = 3.96DD434 pKa = 4.22SEE436 pKa = 4.53EE437 pKa = 4.55IEE439 pKa = 4.88DD440 pKa = 4.14SYY442 pKa = 12.12EE443 pKa = 3.87EE444 pKa = 4.71DD445 pKa = 3.94SEE447 pKa = 5.05DD448 pKa = 4.76DD449 pKa = 3.59EE450 pKa = 5.93SEE452 pKa = 4.41EE453 pKa = 4.04EE454 pKa = 4.17DD455 pKa = 4.25SEE457 pKa = 5.0EE458 pKa = 4.01EE459 pKa = 4.24DD460 pKa = 3.81SEE462 pKa = 4.15EE463 pKa = 4.42DD464 pKa = 3.67EE465 pKa = 4.42EE466 pKa = 5.6SEE468 pKa = 4.71EE469 pKa = 4.11EE470 pKa = 4.28DD471 pKa = 4.15SEE473 pKa = 4.34EE474 pKa = 4.33TEE476 pKa = 4.49DD477 pKa = 4.79SYY479 pKa = 12.2EE480 pKa = 4.03EE481 pKa = 4.51DD482 pKa = 3.6SEE484 pKa = 4.27EE485 pKa = 4.74DD486 pKa = 3.34EE487 pKa = 5.18SEE489 pKa = 4.39EE490 pKa = 3.93EE491 pKa = 4.07DD492 pKa = 3.86SEE494 pKa = 4.61EE495 pKa = 4.28EE496 pKa = 3.92EE497 pKa = 4.8SEE499 pKa = 4.25AEE501 pKa = 3.96PEE503 pKa = 4.14EE504 pKa = 4.39EE505 pKa = 4.61SEE507 pKa = 4.06EE508 pKa = 4.39DD509 pKa = 3.55YY510 pKa = 11.88GDD512 pKa = 4.47AEE514 pKa = 4.01ASYY517 pKa = 11.48DD518 pKa = 3.94EE519 pKa = 4.89GGEE522 pKa = 4.13DD523 pKa = 3.25EE524 pKa = 5.32GGGEE528 pKa = 4.39GEE530 pKa = 4.44EE531 pKa = 4.37EE532 pKa = 4.02
Molecular weight: 60.29 kDa
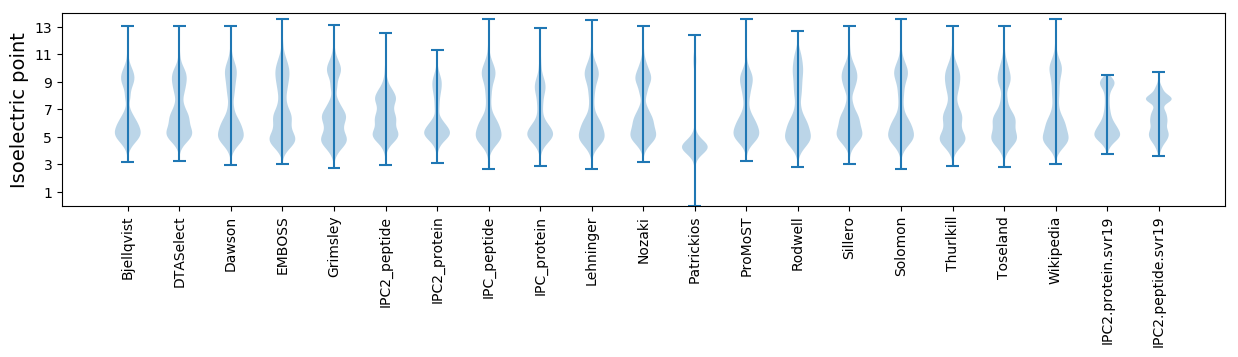
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T1DZU7|A0A2T1DZU7_9CYAN Tfp pilus assembly protein PilF OS=Stenomitos frigidus ULC18 OX=2107698 GN=C7B82_21165 PE=4 SV=1
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 9.47RR13 pKa = 11.84KK14 pKa = 7.16RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TATGRR29 pKa = 11.84RR30 pKa = 11.84VIQARR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.62GRR40 pKa = 11.84FRR42 pKa = 11.84LAVAA46 pKa = 4.63
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 9.47RR13 pKa = 11.84KK14 pKa = 7.16RR15 pKa = 11.84TSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TATGRR29 pKa = 11.84RR30 pKa = 11.84VIQARR35 pKa = 11.84RR36 pKa = 11.84TKK38 pKa = 10.62GRR40 pKa = 11.84FRR42 pKa = 11.84LAVAA46 pKa = 4.63
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1851076 |
25 |
2980 |
312.9 |
34.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.776 ± 0.033 | 0.957 ± 0.01 |
5.146 ± 0.022 | 5.649 ± 0.034 |
3.699 ± 0.021 | 6.81 ± 0.037 |
2.059 ± 0.019 | 5.457 ± 0.023 |
3.867 ± 0.027 | 11.448 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.888 ± 0.014 | 3.36 ± 0.026 |
5.128 ± 0.026 | 5.682 ± 0.037 |
5.736 ± 0.026 | 6.224 ± 0.029 |
6.055 ± 0.032 | 6.872 ± 0.028 |
1.463 ± 0.015 | 2.724 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
