
Pseudovibrio axinellae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Stappiaceae; Pseudovibrio
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
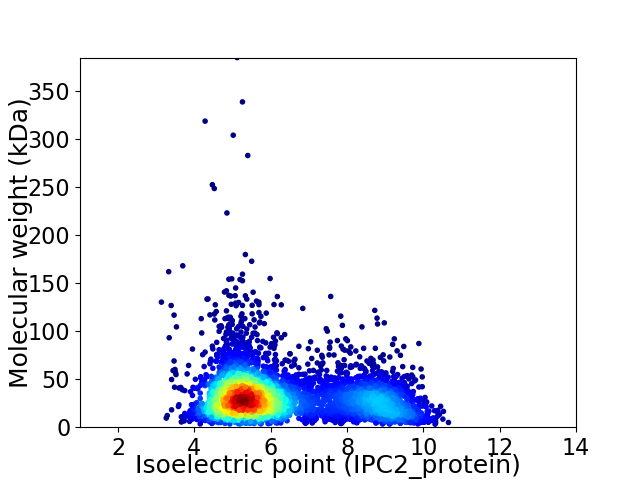
Virtual 2D-PAGE plot for 4634 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A165WXM8|A0A165WXM8_9RHOB Type I secretion system ATP-binding protein PrsD OS=Pseudovibrio axinellae OX=989403 GN=prsD_4 PE=4 SV=1
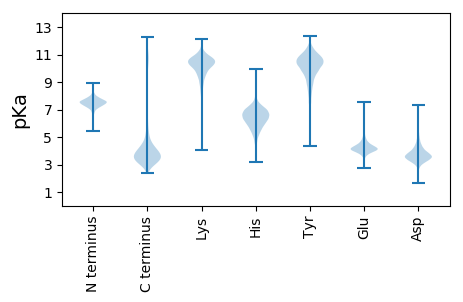
MM1 pKa = 7.5NIKK4 pKa = 8.67TLALAASAAAFATSAQAADD23 pKa = 4.16LPMAAEE29 pKa = 3.99PVDD32 pKa = 3.81YY33 pKa = 11.22VKK35 pKa = 11.11ACDD38 pKa = 3.72AFGAGYY44 pKa = 9.0FQLPGKK50 pKa = 7.03EE51 pKa = 4.1TCIKK55 pKa = 10.23LGGRR59 pKa = 11.84IRR61 pKa = 11.84AQYY64 pKa = 10.59VSQDD68 pKa = 3.66LTDD71 pKa = 3.76KK72 pKa = 10.84STDD75 pKa = 3.33NDD77 pKa = 3.36ATSYY81 pKa = 10.61ARR83 pKa = 11.84GYY85 pKa = 10.49IYY87 pKa = 10.25FNSMTATDD95 pKa = 4.6FGTIKK100 pKa = 10.02TFVEE104 pKa = 5.69LEE106 pKa = 4.31SEE108 pKa = 4.29WNQDD112 pKa = 3.96AEE114 pKa = 4.76GADD117 pKa = 3.57TKK119 pKa = 11.74ANDD122 pKa = 2.79VWIQLGTGYY131 pKa = 11.14GSFLFGRR138 pKa = 11.84EE139 pKa = 3.5ASAFDD144 pKa = 3.96AFTGYY149 pKa = 7.61TWIGPVGNAYY159 pKa = 10.76SDD161 pKa = 3.54TSTLQASFTADD172 pKa = 3.17LGNGLTATASVEE184 pKa = 3.92DD185 pKa = 3.56SAYY188 pKa = 10.6RR189 pKa = 11.84AGDD192 pKa = 3.37NDD194 pKa = 3.64AVDD197 pKa = 4.76FVGALNLSQGWGSFKK212 pKa = 10.59LAAAAHH218 pKa = 5.22NTANSEE224 pKa = 4.2DD225 pKa = 3.59YY226 pKa = 10.68GYY228 pKa = 11.47AVGATAIFNLDD239 pKa = 3.29MVKK242 pKa = 10.53EE243 pKa = 4.31GTEE246 pKa = 3.93VTFQAQYY253 pKa = 11.7ADD255 pKa = 3.69DD256 pKa = 4.42AGNYY260 pKa = 9.36IGIDD264 pKa = 3.46TDD266 pKa = 3.68EE267 pKa = 6.08DD268 pKa = 3.67DD269 pKa = 3.59VDD271 pKa = 3.7AVRR274 pKa = 11.84GYY276 pKa = 10.59SLSAGLEE283 pKa = 3.91TALTDD288 pKa = 3.97KK289 pKa = 11.18VSAQLDD295 pKa = 3.99LSYY298 pKa = 10.56MDD300 pKa = 4.08IEE302 pKa = 4.66STVSGSDD309 pKa = 3.25EE310 pKa = 4.91DD311 pKa = 3.95EE312 pKa = 3.55QTYY315 pKa = 10.62AVNGSVVYY323 pKa = 10.68SPVEE327 pKa = 3.91GLGLALAAGWSDD339 pKa = 3.7GEE341 pKa = 4.07EE342 pKa = 4.26DD343 pKa = 5.56GVDD346 pKa = 3.41KK347 pKa = 11.16DD348 pKa = 5.33DD349 pKa = 3.88EE350 pKa = 4.88VKK352 pKa = 10.86VGARR356 pKa = 11.84VQYY359 pKa = 9.56TFF361 pKa = 4.12
MM1 pKa = 7.5NIKK4 pKa = 8.67TLALAASAAAFATSAQAADD23 pKa = 4.16LPMAAEE29 pKa = 3.99PVDD32 pKa = 3.81YY33 pKa = 11.22VKK35 pKa = 11.11ACDD38 pKa = 3.72AFGAGYY44 pKa = 9.0FQLPGKK50 pKa = 7.03EE51 pKa = 4.1TCIKK55 pKa = 10.23LGGRR59 pKa = 11.84IRR61 pKa = 11.84AQYY64 pKa = 10.59VSQDD68 pKa = 3.66LTDD71 pKa = 3.76KK72 pKa = 10.84STDD75 pKa = 3.33NDD77 pKa = 3.36ATSYY81 pKa = 10.61ARR83 pKa = 11.84GYY85 pKa = 10.49IYY87 pKa = 10.25FNSMTATDD95 pKa = 4.6FGTIKK100 pKa = 10.02TFVEE104 pKa = 5.69LEE106 pKa = 4.31SEE108 pKa = 4.29WNQDD112 pKa = 3.96AEE114 pKa = 4.76GADD117 pKa = 3.57TKK119 pKa = 11.74ANDD122 pKa = 2.79VWIQLGTGYY131 pKa = 11.14GSFLFGRR138 pKa = 11.84EE139 pKa = 3.5ASAFDD144 pKa = 3.96AFTGYY149 pKa = 7.61TWIGPVGNAYY159 pKa = 10.76SDD161 pKa = 3.54TSTLQASFTADD172 pKa = 3.17LGNGLTATASVEE184 pKa = 3.92DD185 pKa = 3.56SAYY188 pKa = 10.6RR189 pKa = 11.84AGDD192 pKa = 3.37NDD194 pKa = 3.64AVDD197 pKa = 4.76FVGALNLSQGWGSFKK212 pKa = 10.59LAAAAHH218 pKa = 5.22NTANSEE224 pKa = 4.2DD225 pKa = 3.59YY226 pKa = 10.68GYY228 pKa = 11.47AVGATAIFNLDD239 pKa = 3.29MVKK242 pKa = 10.53EE243 pKa = 4.31GTEE246 pKa = 3.93VTFQAQYY253 pKa = 11.7ADD255 pKa = 3.69DD256 pKa = 4.42AGNYY260 pKa = 9.36IGIDD264 pKa = 3.46TDD266 pKa = 3.68EE267 pKa = 6.08DD268 pKa = 3.67DD269 pKa = 3.59VDD271 pKa = 3.7AVRR274 pKa = 11.84GYY276 pKa = 10.59SLSAGLEE283 pKa = 3.91TALTDD288 pKa = 3.97KK289 pKa = 11.18VSAQLDD295 pKa = 3.99LSYY298 pKa = 10.56MDD300 pKa = 4.08IEE302 pKa = 4.66STVSGSDD309 pKa = 3.25EE310 pKa = 4.91DD311 pKa = 3.95EE312 pKa = 3.55QTYY315 pKa = 10.62AVNGSVVYY323 pKa = 10.68SPVEE327 pKa = 3.91GLGLALAAGWSDD339 pKa = 3.7GEE341 pKa = 4.07EE342 pKa = 4.26DD343 pKa = 5.56GVDD346 pKa = 3.41KK347 pKa = 11.16DD348 pKa = 5.33DD349 pKa = 3.88EE350 pKa = 4.88VKK352 pKa = 10.86VGARR356 pKa = 11.84VQYY359 pKa = 9.56TFF361 pKa = 4.12
Molecular weight: 38.14 kDa
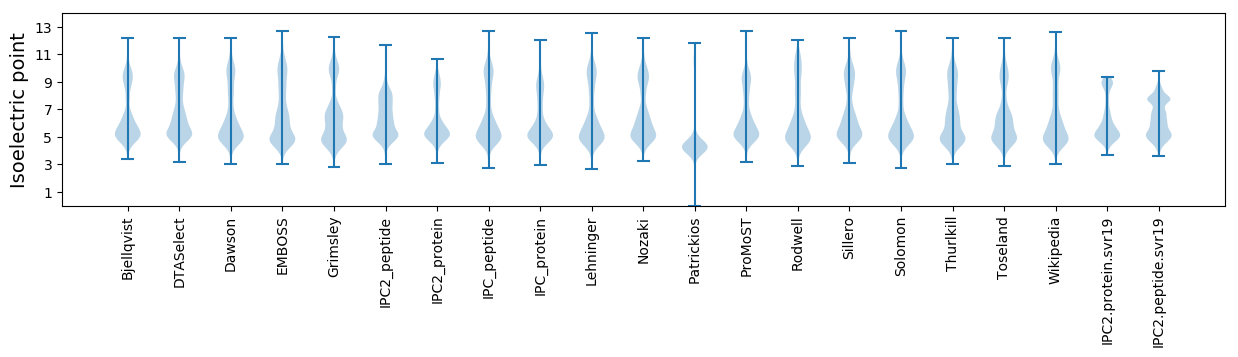
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A165ULJ6|A0A165ULJ6_9RHOB Nucleoside diphosphate kinase OS=Pseudovibrio axinellae OX=989403 GN=ndk PE=3 SV=1
MM1 pKa = 7.44LRR3 pKa = 11.84TATLQLMLSSRR14 pKa = 11.84NSRR17 pKa = 11.84EE18 pKa = 3.83RR19 pKa = 11.84ANGAVLPEE27 pKa = 3.85YY28 pKa = 8.98RR29 pKa = 11.84TLSNCVRR36 pKa = 11.84LFTRR40 pKa = 11.84QAQMTAGQRR49 pKa = 11.84FLEE52 pKa = 4.48SLLRR56 pKa = 11.84WKK58 pKa = 9.6STVSAMAKK66 pKa = 9.59YY67 pKa = 9.02VYY69 pKa = 10.5CMKK72 pKa = 10.79CEE74 pKa = 3.73DD75 pKa = 4.22AIRR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84SLTVCCVACAKK91 pKa = 10.41AAISSTVPTMFLRR104 pKa = 11.84ALGFPLRR111 pKa = 11.84TTHH114 pKa = 7.37CSGSPLKK121 pKa = 10.47KK122 pKa = 9.95QAATTIAPRR131 pKa = 11.84PLLQCWDD138 pKa = 3.27QRR140 pKa = 11.84LLVGG144 pKa = 4.83
MM1 pKa = 7.44LRR3 pKa = 11.84TATLQLMLSSRR14 pKa = 11.84NSRR17 pKa = 11.84EE18 pKa = 3.83RR19 pKa = 11.84ANGAVLPEE27 pKa = 3.85YY28 pKa = 8.98RR29 pKa = 11.84TLSNCVRR36 pKa = 11.84LFTRR40 pKa = 11.84QAQMTAGQRR49 pKa = 11.84FLEE52 pKa = 4.48SLLRR56 pKa = 11.84WKK58 pKa = 9.6STVSAMAKK66 pKa = 9.59YY67 pKa = 9.02VYY69 pKa = 10.5CMKK72 pKa = 10.79CEE74 pKa = 3.73DD75 pKa = 4.22AIRR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84SLTVCCVACAKK91 pKa = 10.41AAISSTVPTMFLRR104 pKa = 11.84ALGFPLRR111 pKa = 11.84TTHH114 pKa = 7.37CSGSPLKK121 pKa = 10.47KK122 pKa = 9.95QAATTIAPRR131 pKa = 11.84PLLQCWDD138 pKa = 3.27QRR140 pKa = 11.84LLVGG144 pKa = 4.83
Molecular weight: 16.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1443537 |
29 |
3466 |
311.5 |
34.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.171 ± 0.041 | 1.006 ± 0.014 |
5.463 ± 0.032 | 6.339 ± 0.037 |
4.107 ± 0.029 | 7.819 ± 0.038 |
2.093 ± 0.02 | 5.705 ± 0.03 |
4.59 ± 0.033 | 10.196 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.587 ± 0.019 | 3.368 ± 0.022 |
4.513 ± 0.023 | 3.703 ± 0.024 |
5.505 ± 0.037 | 6.422 ± 0.031 |
5.405 ± 0.03 | 7.175 ± 0.027 |
1.277 ± 0.014 | 2.558 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
