
Faeces associated gemycircularvirus 17
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus chicas2; Chicken associated gemycircularvirus 2
Average proteome isoelectric point is 7.32
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
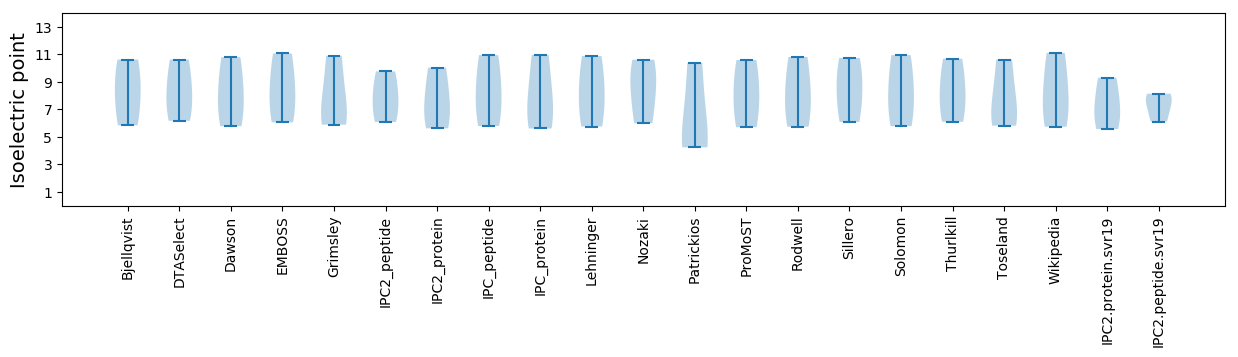
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A160HWK6|A0A160HWK6_9VIRU Capsid protein OS=Faeces associated gemycircularvirus 17 OX=1843737 PE=4 SV=1
MM1 pKa = 7.81PATSLLHH8 pKa = 5.51TRR10 pKa = 11.84SAVLLMSGPLTTILDD25 pKa = 3.9LLEE28 pKa = 5.63RR29 pKa = 11.84SVSWDD34 pKa = 3.3EE35 pKa = 4.64KK36 pKa = 10.16FMLMEE41 pKa = 4.04VLISMFSAISAEE53 pKa = 4.18SFDD56 pKa = 3.92PDD58 pKa = 3.41EE59 pKa = 5.69PISLMCRR66 pKa = 11.84VITQTSSDD74 pKa = 3.44LKK76 pKa = 9.38EE77 pKa = 3.83TLEE80 pKa = 4.21RR81 pKa = 11.84VRR83 pKa = 11.84VTQQKK88 pKa = 8.83MEE90 pKa = 4.28TLSPVGSPWNPCPLRR105 pKa = 11.84SFQALKK111 pKa = 10.52IRR113 pKa = 11.84GLRR116 pKa = 11.84SSAAKK121 pKa = 10.15VEE123 pKa = 4.38RR124 pKa = 11.84NFMRR128 pKa = 11.84LLTTSVRR135 pKa = 11.84GISSPDD141 pKa = 3.12LEE143 pKa = 5.4AFTHH147 pKa = 5.78TPSGSGLRR155 pKa = 11.84SAWATNVPPGSCLAMEE171 pKa = 6.05LFLTWLATEE180 pKa = 4.46RR181 pKa = 11.84EE182 pKa = 3.81FWMYY186 pKa = 10.13EE187 pKa = 3.59VSGSFGRR194 pKa = 11.84VKK196 pKa = 10.09TLVIYY201 pKa = 9.06GPPLTGKK208 pKa = 6.25TTWAQSLGNHH218 pKa = 5.91VYY220 pKa = 9.37MKK222 pKa = 9.72KK223 pKa = 8.63TFNAKK228 pKa = 9.31EE229 pKa = 3.8ASLCEE234 pKa = 4.24SKK236 pKa = 10.65DD237 pKa = 3.31YY238 pKa = 11.35CVIDD242 pKa = 5.38DD243 pKa = 3.72ISGGIKK249 pKa = 10.12FFPHH253 pKa = 5.15WKK255 pKa = 9.73DD256 pKa = 2.82WFGGQPYY263 pKa = 9.56VQIRR267 pKa = 11.84LLYY270 pKa = 10.03RR271 pKa = 11.84DD272 pKa = 3.99EE273 pKa = 5.47LLLRR277 pKa = 11.84WGKK280 pKa = 7.53PTIWLNNRR288 pKa = 11.84DD289 pKa = 3.78PRR291 pKa = 11.84DD292 pKa = 3.39QLRR295 pKa = 11.84DD296 pKa = 3.56MVGRR300 pKa = 11.84DD301 pKa = 3.72YY302 pKa = 11.65SDD304 pKa = 3.39EE305 pKa = 4.01QYY307 pKa = 11.48QNDD310 pKa = 4.55VEE312 pKa = 4.8WMDD315 pKa = 3.81ANCIFVYY322 pKa = 10.21VDD324 pKa = 3.3SPIVTFHH331 pKa = 7.16ANTEE335 pKa = 4.1
MM1 pKa = 7.81PATSLLHH8 pKa = 5.51TRR10 pKa = 11.84SAVLLMSGPLTTILDD25 pKa = 3.9LLEE28 pKa = 5.63RR29 pKa = 11.84SVSWDD34 pKa = 3.3EE35 pKa = 4.64KK36 pKa = 10.16FMLMEE41 pKa = 4.04VLISMFSAISAEE53 pKa = 4.18SFDD56 pKa = 3.92PDD58 pKa = 3.41EE59 pKa = 5.69PISLMCRR66 pKa = 11.84VITQTSSDD74 pKa = 3.44LKK76 pKa = 9.38EE77 pKa = 3.83TLEE80 pKa = 4.21RR81 pKa = 11.84VRR83 pKa = 11.84VTQQKK88 pKa = 8.83MEE90 pKa = 4.28TLSPVGSPWNPCPLRR105 pKa = 11.84SFQALKK111 pKa = 10.52IRR113 pKa = 11.84GLRR116 pKa = 11.84SSAAKK121 pKa = 10.15VEE123 pKa = 4.38RR124 pKa = 11.84NFMRR128 pKa = 11.84LLTTSVRR135 pKa = 11.84GISSPDD141 pKa = 3.12LEE143 pKa = 5.4AFTHH147 pKa = 5.78TPSGSGLRR155 pKa = 11.84SAWATNVPPGSCLAMEE171 pKa = 6.05LFLTWLATEE180 pKa = 4.46RR181 pKa = 11.84EE182 pKa = 3.81FWMYY186 pKa = 10.13EE187 pKa = 3.59VSGSFGRR194 pKa = 11.84VKK196 pKa = 10.09TLVIYY201 pKa = 9.06GPPLTGKK208 pKa = 6.25TTWAQSLGNHH218 pKa = 5.91VYY220 pKa = 9.37MKK222 pKa = 9.72KK223 pKa = 8.63TFNAKK228 pKa = 9.31EE229 pKa = 3.8ASLCEE234 pKa = 4.24SKK236 pKa = 10.65DD237 pKa = 3.31YY238 pKa = 11.35CVIDD242 pKa = 5.38DD243 pKa = 3.72ISGGIKK249 pKa = 10.12FFPHH253 pKa = 5.15WKK255 pKa = 9.73DD256 pKa = 2.82WFGGQPYY263 pKa = 9.56VQIRR267 pKa = 11.84LLYY270 pKa = 10.03RR271 pKa = 11.84DD272 pKa = 3.99EE273 pKa = 5.47LLLRR277 pKa = 11.84WGKK280 pKa = 7.53PTIWLNNRR288 pKa = 11.84DD289 pKa = 3.78PRR291 pKa = 11.84DD292 pKa = 3.39QLRR295 pKa = 11.84DD296 pKa = 3.56MVGRR300 pKa = 11.84DD301 pKa = 3.72YY302 pKa = 11.65SDD304 pKa = 3.39EE305 pKa = 4.01QYY307 pKa = 11.48QNDD310 pKa = 4.55VEE312 pKa = 4.8WMDD315 pKa = 3.81ANCIFVYY322 pKa = 10.21VDD324 pKa = 3.3SPIVTFHH331 pKa = 7.16ANTEE335 pKa = 4.1
Molecular weight: 38.16 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A168MG03|A0A168MG03_9VIRU RepA OS=Faeces associated gemycircularvirus 17 OX=1843737 PE=4 SV=1
MM1 pKa = 7.91PYY3 pKa = 10.17QRR5 pKa = 11.84RR6 pKa = 11.84SYY8 pKa = 10.42SSRR11 pKa = 11.84ARR13 pKa = 11.84RR14 pKa = 11.84PTYY17 pKa = 9.22RR18 pKa = 11.84TARR21 pKa = 11.84PARR24 pKa = 11.84YY25 pKa = 7.18TRR27 pKa = 11.84NSYY30 pKa = 8.89SRR32 pKa = 11.84RR33 pKa = 11.84PTSRR37 pKa = 11.84RR38 pKa = 11.84TPIRR42 pKa = 11.84RR43 pKa = 11.84TYY45 pKa = 10.43RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84STEE51 pKa = 3.11KK52 pKa = 7.8MPRR55 pKa = 11.84YY56 pKa = 8.51RR57 pKa = 11.84RR58 pKa = 11.84SYY60 pKa = 9.28RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84PMTRR68 pKa = 11.84KK69 pKa = 10.06KK70 pKa = 10.2ILNLTSVKK78 pKa = 10.53KK79 pKa = 9.9QDD81 pKa = 3.56NMQLSSNVTAATPQGSATYY100 pKa = 8.61TNSAAVLPATQLYY113 pKa = 8.64IFPWIATARR122 pKa = 11.84DD123 pKa = 3.37LDD125 pKa = 4.07RR126 pKa = 11.84DD127 pKa = 3.64TGNIRR132 pKa = 11.84GAPSDD137 pKa = 3.69SATRR141 pKa = 11.84TAQTCYY147 pKa = 9.91MVGLRR152 pKa = 11.84EE153 pKa = 4.26NIEE156 pKa = 3.65ITTNDD161 pKa = 3.41GLPWQWRR168 pKa = 11.84RR169 pKa = 11.84ICFTFKK175 pKa = 10.65GSEE178 pKa = 4.09FTATSTTGLNYY189 pKa = 10.39YY190 pKa = 9.97LQTSNGMTRR199 pKa = 11.84TVNTAYY205 pKa = 10.44GSTAVGVLQSLLFKK219 pKa = 10.76GAAGVDD225 pKa = 3.33WSNTITAPTDD235 pKa = 3.09NTTISVKK242 pKa = 9.75YY243 pKa = 10.31DD244 pKa = 3.24RR245 pKa = 11.84TISIASGNEE254 pKa = 3.19QGTIRR259 pKa = 11.84RR260 pKa = 11.84YY261 pKa = 10.17RR262 pKa = 11.84RR263 pKa = 11.84FHH265 pKa = 6.47PMGHH269 pKa = 6.24NIIYY273 pKa = 10.52ADD275 pKa = 4.11DD276 pKa = 3.96EE277 pKa = 4.51QGGTEE282 pKa = 4.07SEE284 pKa = 4.59SFVSSRR290 pKa = 11.84GKK292 pKa = 9.8PGMGDD297 pKa = 3.54YY298 pKa = 10.92YY299 pKa = 11.12VIDD302 pKa = 3.42IFQPRR307 pKa = 11.84FGGTSASQLRR317 pKa = 11.84FEE319 pKa = 5.14PSATLYY325 pKa = 9.32WHH327 pKa = 6.99EE328 pKa = 4.35KK329 pKa = 9.3
MM1 pKa = 7.91PYY3 pKa = 10.17QRR5 pKa = 11.84RR6 pKa = 11.84SYY8 pKa = 10.42SSRR11 pKa = 11.84ARR13 pKa = 11.84RR14 pKa = 11.84PTYY17 pKa = 9.22RR18 pKa = 11.84TARR21 pKa = 11.84PARR24 pKa = 11.84YY25 pKa = 7.18TRR27 pKa = 11.84NSYY30 pKa = 8.89SRR32 pKa = 11.84RR33 pKa = 11.84PTSRR37 pKa = 11.84RR38 pKa = 11.84TPIRR42 pKa = 11.84RR43 pKa = 11.84TYY45 pKa = 10.43RR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84STEE51 pKa = 3.11KK52 pKa = 7.8MPRR55 pKa = 11.84YY56 pKa = 8.51RR57 pKa = 11.84RR58 pKa = 11.84SYY60 pKa = 9.28RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84RR64 pKa = 11.84PMTRR68 pKa = 11.84KK69 pKa = 10.06KK70 pKa = 10.2ILNLTSVKK78 pKa = 10.53KK79 pKa = 9.9QDD81 pKa = 3.56NMQLSSNVTAATPQGSATYY100 pKa = 8.61TNSAAVLPATQLYY113 pKa = 8.64IFPWIATARR122 pKa = 11.84DD123 pKa = 3.37LDD125 pKa = 4.07RR126 pKa = 11.84DD127 pKa = 3.64TGNIRR132 pKa = 11.84GAPSDD137 pKa = 3.69SATRR141 pKa = 11.84TAQTCYY147 pKa = 9.91MVGLRR152 pKa = 11.84EE153 pKa = 4.26NIEE156 pKa = 3.65ITTNDD161 pKa = 3.41GLPWQWRR168 pKa = 11.84RR169 pKa = 11.84ICFTFKK175 pKa = 10.65GSEE178 pKa = 4.09FTATSTTGLNYY189 pKa = 10.39YY190 pKa = 9.97LQTSNGMTRR199 pKa = 11.84TVNTAYY205 pKa = 10.44GSTAVGVLQSLLFKK219 pKa = 10.76GAAGVDD225 pKa = 3.33WSNTITAPTDD235 pKa = 3.09NTTISVKK242 pKa = 9.75YY243 pKa = 10.31DD244 pKa = 3.24RR245 pKa = 11.84TISIASGNEE254 pKa = 3.19QGTIRR259 pKa = 11.84RR260 pKa = 11.84YY261 pKa = 10.17RR262 pKa = 11.84RR263 pKa = 11.84FHH265 pKa = 6.47PMGHH269 pKa = 6.24NIIYY273 pKa = 10.52ADD275 pKa = 4.11DD276 pKa = 3.96EE277 pKa = 4.51QGGTEE282 pKa = 4.07SEE284 pKa = 4.59SFVSSRR290 pKa = 11.84GKK292 pKa = 9.8PGMGDD297 pKa = 3.54YY298 pKa = 10.92YY299 pKa = 11.12VIDD302 pKa = 3.42IFQPRR307 pKa = 11.84FGGTSASQLRR317 pKa = 11.84FEE319 pKa = 5.14PSATLYY325 pKa = 9.32WHH327 pKa = 6.99EE328 pKa = 4.35KK329 pKa = 9.3
Molecular weight: 37.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
916 |
252 |
335 |
305.3 |
34.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.441 ± 0.623 | 1.419 ± 0.36 |
4.258 ± 0.669 | 5.022 ± 0.879 |
3.712 ± 0.376 | 6.004 ± 0.341 |
1.31 ± 0.177 | 4.476 ± 0.351 |
3.275 ± 0.422 | 9.061 ± 2.011 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.384 ± 0.423 | 3.275 ± 0.594 |
5.895 ± 0.438 | 2.948 ± 0.519 |
8.952 ± 1.748 | 10.917 ± 1.263 |
9.061 ± 1.514 | 4.803 ± 0.759 |
2.511 ± 0.466 | 3.275 ± 1.303 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
