
Photobacterium damselae subsp. damselae CIP 102761
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Vibrionales; Vibrionaceae; Photobacterium; Photobacterium damselae; Photobacterium damselae subsp. damselae
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
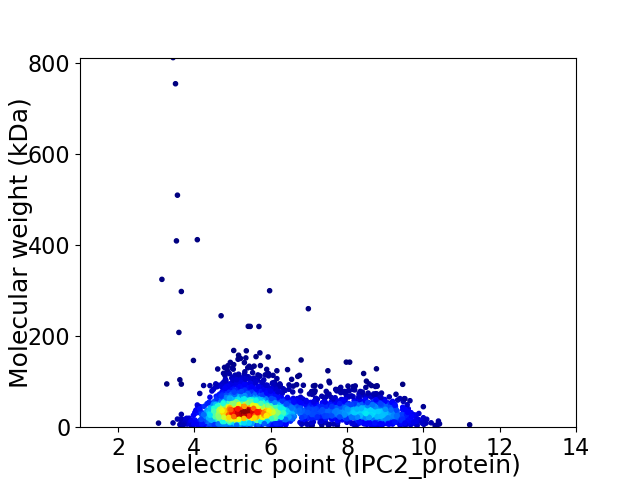
Virtual 2D-PAGE plot for 3498 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D0Z4Q9|D0Z4Q9_PHODD Inosine-uridine preferring nucleoside hydrolase OS=Photobacterium damselae subsp. damselae CIP 102761 OX=675817 GN=VDA_000517 PE=4 SV=1
MM1 pKa = 7.31NVFKK5 pKa = 11.07GIAKK9 pKa = 10.21VGITAVASISFAAMALTEE27 pKa = 4.39SGTVIKK33 pKa = 10.3NQAGATYY40 pKa = 9.91RR41 pKa = 11.84DD42 pKa = 3.47SANILRR48 pKa = 11.84STTSNLVEE56 pKa = 4.21TLVQPVAALTLINDD70 pKa = 3.68QSKK73 pKa = 10.8LGAPGRR79 pKa = 11.84TVTFNHH85 pKa = 5.99VLTNTGNSTDD95 pKa = 3.66SFALDD100 pKa = 3.4MVLPASVISSATIYY114 pKa = 10.49PDD116 pKa = 3.44TNNDD120 pKa = 2.91GVADD124 pKa = 4.41PGSTPIASGDD134 pKa = 3.64VVGPLAPGEE143 pKa = 4.19SFEE146 pKa = 4.27FVIVADD152 pKa = 4.2IDD154 pKa = 3.62PAATINSSNALTITATSQAVVVDD177 pKa = 4.86GVAVANGGVSAQTNTDD193 pKa = 3.0TVTATNLPIVEE204 pKa = 4.09ITKK207 pKa = 10.5ALSSSEE213 pKa = 4.07GASPSGPYY221 pKa = 9.1TVTFTYY227 pKa = 10.68TNVGLTDD234 pKa = 3.89MDD236 pKa = 4.33PLQTNGVILNDD247 pKa = 3.52VLPEE251 pKa = 3.81GMRR254 pKa = 11.84FNGAVSWSISGTTITASGTTTGGSEE279 pKa = 4.05TSGDD283 pKa = 3.15QDD285 pKa = 4.74LAFTTCIAPDD295 pKa = 4.01ASCPNNDD302 pKa = 3.58EE303 pKa = 4.22IQFVMDD309 pKa = 4.52GLASDD314 pKa = 4.65EE315 pKa = 4.29SATISFEE322 pKa = 4.24VNIDD326 pKa = 3.56SGLSATTLYY335 pKa = 11.01NQGTYY340 pKa = 10.98GFDD343 pKa = 3.45EE344 pKa = 5.2DD345 pKa = 4.25NSGTVDD351 pKa = 3.54VGEE354 pKa = 4.0QDD356 pKa = 3.12STNTNRR362 pKa = 11.84VPFLITSNYY371 pKa = 9.76AVIANNGGCDD381 pKa = 2.94VGTDD385 pKa = 3.88NNCDD389 pKa = 3.33GSDD392 pKa = 3.79DD393 pKa = 4.05TNHH396 pKa = 6.92EE397 pKa = 4.13IVDD400 pKa = 3.73IASASQGARR409 pKa = 11.84VSFTNYY415 pKa = 8.22IWNTGNDD422 pKa = 3.08EE423 pKa = 5.01DD424 pKa = 4.64IFNISIEE431 pKa = 4.22SSTFPSGTSFFLYY444 pKa = 10.26KK445 pKa = 10.15SDD447 pKa = 3.82GVTPLIDD454 pKa = 3.68TDD456 pKa = 3.7NDD458 pKa = 4.22SNPDD462 pKa = 3.35TGVIPAAGEE471 pKa = 4.09TCPSYY476 pKa = 11.15LVTDD480 pKa = 4.81GNLCGYY486 pKa = 10.1KK487 pKa = 9.96VVLVATLPASALGGPYY503 pKa = 10.12QITKK507 pKa = 10.21RR508 pKa = 11.84ATSSSDD514 pKa = 2.93TSKK517 pKa = 10.1TNTVIDD523 pKa = 3.74RR524 pKa = 11.84LGTITASTVDD534 pKa = 3.35LTNNFRR540 pKa = 11.84AEE542 pKa = 3.9TSLTVEE548 pKa = 4.85DD549 pKa = 4.39SCDD552 pKa = 3.72AADD555 pKa = 4.34DD556 pKa = 3.94NCGIGAGPEE565 pKa = 4.04ATPVTTNTTAPGTTTRR581 pKa = 11.84FTLYY585 pKa = 8.97VTNTSGAADD594 pKa = 3.86SYY596 pKa = 10.2EE597 pKa = 3.95LQYY600 pKa = 11.45SSTNFAEE607 pKa = 4.24NTMPADD613 pKa = 3.14WSVVFKK619 pKa = 10.96NAAGEE624 pKa = 4.36VITNIPVLGPDD635 pKa = 3.78DD636 pKa = 4.84AMLVYY641 pKa = 10.93ADD643 pKa = 3.4VTVPEE648 pKa = 4.3NATTATQDD656 pKa = 3.13VYY658 pKa = 11.22FKK660 pKa = 10.91VVSPTTGASDD670 pKa = 3.63IKK672 pKa = 10.59RR673 pKa = 11.84DD674 pKa = 3.47AVTIDD679 pKa = 3.73NSSQCLAFGPPNANGVTYY697 pKa = 10.58AGGSIVYY704 pKa = 9.34KK705 pKa = 10.52HH706 pKa = 6.25ILNNTSNDD714 pKa = 3.11AYY716 pKa = 10.04TNIALTVTNSDD727 pKa = 3.06SDD729 pKa = 3.86FSAVIYY735 pKa = 10.52EE736 pKa = 4.37DD737 pKa = 3.79TNGDD741 pKa = 3.84DD742 pKa = 3.98VLTGADD748 pKa = 3.48NSITSIASLAGGAQKK763 pKa = 11.08VLFVKK768 pKa = 10.47VLAPANASQGQDD780 pKa = 2.71NITTVEE786 pKa = 4.16TTVACGTLQIIDD798 pKa = 3.8TTTIANTNMKK808 pKa = 8.2VTKK811 pKa = 9.85EE812 pKa = 3.87QTPDD816 pKa = 3.26LDD818 pKa = 4.55CNGTSDD824 pKa = 3.28TGNFVTTNFAVAPGEE839 pKa = 4.19CIIYY843 pKa = 10.66RR844 pKa = 11.84LTTEE848 pKa = 4.57NIAIEE853 pKa = 4.37DD854 pKa = 3.67AHH856 pKa = 6.58NVTLTDD862 pKa = 3.32ATPAYY867 pKa = 6.73TTFNVTGGIPSITEE881 pKa = 3.66GTINTVINGTTGSIIGTVGTVAPGEE906 pKa = 4.32SEE908 pKa = 3.94TLIFGIRR915 pKa = 11.84IDD917 pKa = 3.57NN918 pKa = 3.72
MM1 pKa = 7.31NVFKK5 pKa = 11.07GIAKK9 pKa = 10.21VGITAVASISFAAMALTEE27 pKa = 4.39SGTVIKK33 pKa = 10.3NQAGATYY40 pKa = 9.91RR41 pKa = 11.84DD42 pKa = 3.47SANILRR48 pKa = 11.84STTSNLVEE56 pKa = 4.21TLVQPVAALTLINDD70 pKa = 3.68QSKK73 pKa = 10.8LGAPGRR79 pKa = 11.84TVTFNHH85 pKa = 5.99VLTNTGNSTDD95 pKa = 3.66SFALDD100 pKa = 3.4MVLPASVISSATIYY114 pKa = 10.49PDD116 pKa = 3.44TNNDD120 pKa = 2.91GVADD124 pKa = 4.41PGSTPIASGDD134 pKa = 3.64VVGPLAPGEE143 pKa = 4.19SFEE146 pKa = 4.27FVIVADD152 pKa = 4.2IDD154 pKa = 3.62PAATINSSNALTITATSQAVVVDD177 pKa = 4.86GVAVANGGVSAQTNTDD193 pKa = 3.0TVTATNLPIVEE204 pKa = 4.09ITKK207 pKa = 10.5ALSSSEE213 pKa = 4.07GASPSGPYY221 pKa = 9.1TVTFTYY227 pKa = 10.68TNVGLTDD234 pKa = 3.89MDD236 pKa = 4.33PLQTNGVILNDD247 pKa = 3.52VLPEE251 pKa = 3.81GMRR254 pKa = 11.84FNGAVSWSISGTTITASGTTTGGSEE279 pKa = 4.05TSGDD283 pKa = 3.15QDD285 pKa = 4.74LAFTTCIAPDD295 pKa = 4.01ASCPNNDD302 pKa = 3.58EE303 pKa = 4.22IQFVMDD309 pKa = 4.52GLASDD314 pKa = 4.65EE315 pKa = 4.29SATISFEE322 pKa = 4.24VNIDD326 pKa = 3.56SGLSATTLYY335 pKa = 11.01NQGTYY340 pKa = 10.98GFDD343 pKa = 3.45EE344 pKa = 5.2DD345 pKa = 4.25NSGTVDD351 pKa = 3.54VGEE354 pKa = 4.0QDD356 pKa = 3.12STNTNRR362 pKa = 11.84VPFLITSNYY371 pKa = 9.76AVIANNGGCDD381 pKa = 2.94VGTDD385 pKa = 3.88NNCDD389 pKa = 3.33GSDD392 pKa = 3.79DD393 pKa = 4.05TNHH396 pKa = 6.92EE397 pKa = 4.13IVDD400 pKa = 3.73IASASQGARR409 pKa = 11.84VSFTNYY415 pKa = 8.22IWNTGNDD422 pKa = 3.08EE423 pKa = 5.01DD424 pKa = 4.64IFNISIEE431 pKa = 4.22SSTFPSGTSFFLYY444 pKa = 10.26KK445 pKa = 10.15SDD447 pKa = 3.82GVTPLIDD454 pKa = 3.68TDD456 pKa = 3.7NDD458 pKa = 4.22SNPDD462 pKa = 3.35TGVIPAAGEE471 pKa = 4.09TCPSYY476 pKa = 11.15LVTDD480 pKa = 4.81GNLCGYY486 pKa = 10.1KK487 pKa = 9.96VVLVATLPASALGGPYY503 pKa = 10.12QITKK507 pKa = 10.21RR508 pKa = 11.84ATSSSDD514 pKa = 2.93TSKK517 pKa = 10.1TNTVIDD523 pKa = 3.74RR524 pKa = 11.84LGTITASTVDD534 pKa = 3.35LTNNFRR540 pKa = 11.84AEE542 pKa = 3.9TSLTVEE548 pKa = 4.85DD549 pKa = 4.39SCDD552 pKa = 3.72AADD555 pKa = 4.34DD556 pKa = 3.94NCGIGAGPEE565 pKa = 4.04ATPVTTNTTAPGTTTRR581 pKa = 11.84FTLYY585 pKa = 8.97VTNTSGAADD594 pKa = 3.86SYY596 pKa = 10.2EE597 pKa = 3.95LQYY600 pKa = 11.45SSTNFAEE607 pKa = 4.24NTMPADD613 pKa = 3.14WSVVFKK619 pKa = 10.96NAAGEE624 pKa = 4.36VITNIPVLGPDD635 pKa = 3.78DD636 pKa = 4.84AMLVYY641 pKa = 10.93ADD643 pKa = 3.4VTVPEE648 pKa = 4.3NATTATQDD656 pKa = 3.13VYY658 pKa = 11.22FKK660 pKa = 10.91VVSPTTGASDD670 pKa = 3.63IKK672 pKa = 10.59RR673 pKa = 11.84DD674 pKa = 3.47AVTIDD679 pKa = 3.73NSSQCLAFGPPNANGVTYY697 pKa = 10.58AGGSIVYY704 pKa = 9.34KK705 pKa = 10.52HH706 pKa = 6.25ILNNTSNDD714 pKa = 3.11AYY716 pKa = 10.04TNIALTVTNSDD727 pKa = 3.06SDD729 pKa = 3.86FSAVIYY735 pKa = 10.52EE736 pKa = 4.37DD737 pKa = 3.79TNGDD741 pKa = 3.84DD742 pKa = 3.98VLTGADD748 pKa = 3.48NSITSIASLAGGAQKK763 pKa = 11.08VLFVKK768 pKa = 10.47VLAPANASQGQDD780 pKa = 2.71NITTVEE786 pKa = 4.16TTVACGTLQIIDD798 pKa = 3.8TTTIANTNMKK808 pKa = 8.2VTKK811 pKa = 9.85EE812 pKa = 3.87QTPDD816 pKa = 3.26LDD818 pKa = 4.55CNGTSDD824 pKa = 3.28TGNFVTTNFAVAPGEE839 pKa = 4.19CIIYY843 pKa = 10.66RR844 pKa = 11.84LTTEE848 pKa = 4.57NIAIEE853 pKa = 4.37DD854 pKa = 3.67AHH856 pKa = 6.58NVTLTDD862 pKa = 3.32ATPAYY867 pKa = 6.73TTFNVTGGIPSITEE881 pKa = 3.66GTINTVINGTTGSIIGTVGTVAPGEE906 pKa = 4.32SEE908 pKa = 3.94TLIFGIRR915 pKa = 11.84IDD917 pKa = 3.57NN918 pKa = 3.72
Molecular weight: 94.73 kDa
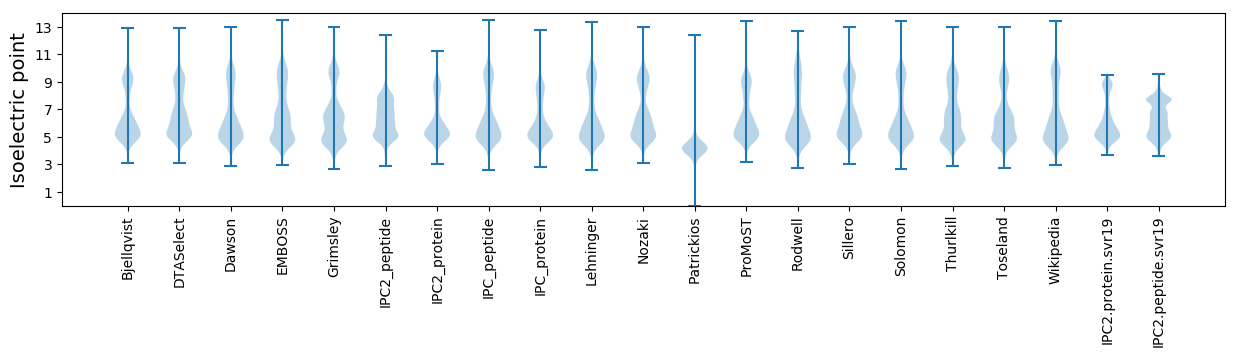
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D0Z141|D0Z141_PHODD GDT1 family protein OS=Photobacterium damselae subsp. damselae CIP 102761 OX=675817 GN=VDA_003255 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPSVLKK12 pKa = 10.49RR13 pKa = 11.84KK14 pKa = 7.97RR15 pKa = 11.84THH17 pKa = 5.89GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.41NGRR29 pKa = 11.84AVINARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.74GRR40 pKa = 11.84KK41 pKa = 8.91RR42 pKa = 11.84LSKK45 pKa = 10.84
MM1 pKa = 7.69SKK3 pKa = 8.98RR4 pKa = 11.84TFQPSVLKK12 pKa = 10.49RR13 pKa = 11.84KK14 pKa = 7.97RR15 pKa = 11.84THH17 pKa = 5.89GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.41NGRR29 pKa = 11.84AVINARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.74GRR40 pKa = 11.84KK41 pKa = 8.91RR42 pKa = 11.84LSKK45 pKa = 10.84
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1276815 |
37 |
7833 |
365.0 |
40.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.464 ± 0.05 | 1.067 ± 0.017 |
5.63 ± 0.07 | 5.927 ± 0.041 |
4.041 ± 0.031 | 6.736 ± 0.062 |
2.311 ± 0.025 | 6.695 ± 0.034 |
5.343 ± 0.061 | 10.196 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.613 ± 0.029 | 4.526 ± 0.04 |
3.985 ± 0.03 | 4.902 ± 0.042 |
4.386 ± 0.043 | 6.385 ± 0.039 |
5.623 ± 0.062 | 6.726 ± 0.043 |
1.24 ± 0.017 | 3.202 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
