
Beihai Nido-like virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Ronidovirineae; Euroniviridae; Ceronivirinae; Charybnivirus; Cradenivirus; Charybnivirus 1
Average proteome isoelectric point is 7.27
Get precalculated fractions of proteins
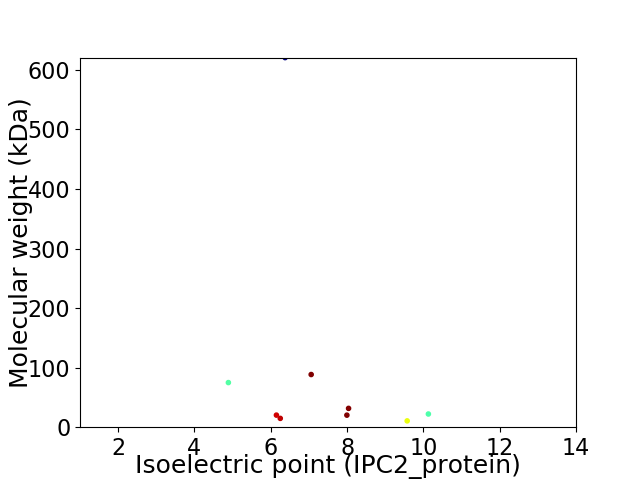
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJ32|A0A1L3KJ32_9NIDO Putative glycoprotein 2 OS=Beihai Nido-like virus 2 OX=1922351 PE=4 SV=1
MM1 pKa = 7.56ALHH4 pKa = 7.13AKK6 pKa = 9.9FLLLVLILLCDD17 pKa = 3.56YY18 pKa = 10.68ARR20 pKa = 11.84SIEE23 pKa = 4.24IVGEE27 pKa = 4.01LRR29 pKa = 11.84NLDD32 pKa = 3.78PGTVKK37 pKa = 10.65GLAARR42 pKa = 11.84GSQLTRR48 pKa = 11.84YY49 pKa = 7.63TNFVQVSRR57 pKa = 11.84PTQGTVRR64 pKa = 11.84EE65 pKa = 4.21DD66 pKa = 3.5HH67 pKa = 6.29YY68 pKa = 11.39VPHH71 pKa = 6.41YY72 pKa = 11.09VDD74 pKa = 4.45IPLDD78 pKa = 3.5TDD80 pKa = 3.65LDD82 pKa = 4.3YY83 pKa = 11.66VVTGTSSLGTDD94 pKa = 4.02FGFRR98 pKa = 11.84VKK100 pKa = 10.65VDD102 pKa = 3.63DD103 pKa = 4.31VNVEE107 pKa = 4.04VEE109 pKa = 4.05NEE111 pKa = 4.38LIYY114 pKa = 9.65TAPFYY119 pKa = 10.71RR120 pKa = 11.84DD121 pKa = 3.06SHH123 pKa = 6.7EE124 pKa = 5.47DD125 pKa = 3.37NLHH128 pKa = 4.98QMVWNWDD135 pKa = 3.58GSSLTHH141 pKa = 6.22GVHH144 pKa = 6.45IPTVCAGLSNVDD156 pKa = 3.46YY157 pKa = 11.53SFFEE161 pKa = 4.41CLRR164 pKa = 11.84TRR166 pKa = 11.84DD167 pKa = 3.48EE168 pKa = 4.3VAQLSQKK175 pKa = 10.54DD176 pKa = 3.82RR177 pKa = 11.84EE178 pKa = 4.39CRR180 pKa = 11.84YY181 pKa = 10.47APLGNEE187 pKa = 3.93YY188 pKa = 10.64LSVGNYY194 pKa = 6.53TVKK197 pKa = 10.63NPRR200 pKa = 11.84GTCLNAEE207 pKa = 4.26SQSWWGRR214 pKa = 11.84PFFTGLYY221 pKa = 6.94KK222 pKa = 10.47TGMPWIRR229 pKa = 11.84AIDD232 pKa = 3.55QWNLADD238 pKa = 5.4CGTGSCTPSGQIRR251 pKa = 11.84VLQRR255 pKa = 11.84KK256 pKa = 7.99WISGVNTAMFFKK268 pKa = 10.31STGKK272 pKa = 10.54AIANYY277 pKa = 8.12TISVSPEE284 pKa = 3.66YY285 pKa = 11.12GCDD288 pKa = 3.6FDD290 pKa = 5.31RR291 pKa = 11.84CEE293 pKa = 4.07LHH295 pKa = 7.43GPGDD299 pKa = 3.67KK300 pKa = 10.0CTLSVAGDD308 pKa = 3.61TMSITGNDD316 pKa = 3.6LEE318 pKa = 4.86TTTRR322 pKa = 11.84MNIMVIAACGDD333 pKa = 3.51YY334 pKa = 10.9SGCQPVAVYY343 pKa = 9.69RR344 pKa = 11.84WPDD347 pKa = 3.43DD348 pKa = 4.35LYY350 pKa = 10.66LTNAFPYY357 pKa = 9.88LKK359 pKa = 9.13WRR361 pKa = 11.84SVPGPVYY368 pKa = 10.77ADD370 pKa = 3.16EE371 pKa = 5.2GKK373 pKa = 8.7TKK375 pKa = 10.36FQADD379 pKa = 3.27AKK381 pKa = 9.95FFPDD385 pKa = 2.81VHH387 pKa = 7.88INEE390 pKa = 4.59EE391 pKa = 4.0FCLDD395 pKa = 3.74LGYY398 pKa = 10.76RR399 pKa = 11.84QDD401 pKa = 3.78EE402 pKa = 5.05GIAQYY407 pKa = 8.32THH409 pKa = 6.75SSNKK413 pKa = 10.55DD414 pKa = 2.59GDD416 pKa = 4.23ANKK419 pKa = 9.45PACRR423 pKa = 11.84KK424 pKa = 8.39KK425 pKa = 10.93APIISQSQRR434 pKa = 11.84EE435 pKa = 4.1FEE437 pKa = 4.13EE438 pKa = 4.62RR439 pKa = 11.84GLSIDD444 pKa = 3.06RR445 pKa = 11.84CGDD448 pKa = 2.75IRR450 pKa = 11.84LEE452 pKa = 4.07YY453 pKa = 10.88VGDD456 pKa = 3.53IDD458 pKa = 5.53MVTLKK463 pKa = 10.67YY464 pKa = 10.94GKK466 pKa = 10.54GYY468 pKa = 10.34DD469 pKa = 3.93GKK471 pKa = 10.66DD472 pKa = 3.31CEE474 pKa = 4.83DD475 pKa = 3.41LKK477 pKa = 11.19KK478 pKa = 11.04VNPHH482 pKa = 6.91PYY484 pKa = 9.66RR485 pKa = 11.84YY486 pKa = 9.24PLSAVKK492 pKa = 9.97AYY494 pKa = 11.25YY495 pKa = 9.83NGCTGGVARR504 pKa = 11.84AYY506 pKa = 10.69FDD508 pKa = 3.32IKK510 pKa = 10.2STEE513 pKa = 4.02IVIDD517 pKa = 4.1PPDD520 pKa = 3.98GVDD523 pKa = 5.49LSVCQIKK530 pKa = 9.38MEE532 pKa = 4.67GYY534 pKa = 9.38YY535 pKa = 11.05GLDD538 pKa = 2.95NGITLTIGGCTVSGPVRR555 pKa = 11.84LQLPQLDD562 pKa = 3.83AYY564 pKa = 10.74FYY566 pKa = 11.15GDD568 pKa = 3.82SKK570 pKa = 10.27TLTTVLGASEE580 pKa = 4.17SRR582 pKa = 11.84ITITMLSYY590 pKa = 11.54YY591 pKa = 10.36DD592 pKa = 3.53VLLIAEE598 pKa = 4.72VFSSTITEE606 pKa = 4.4VPVTALNADD615 pKa = 3.83DD616 pKa = 5.97AYY618 pKa = 9.41TPDD621 pKa = 4.45VIPPPSNDD629 pKa = 2.79GGGFFDD635 pKa = 5.14FLNNMFGNTFSIILIISAVILVGFILVSCCASARR669 pKa = 11.84GGVKK673 pKa = 10.04KK674 pKa = 10.69EE675 pKa = 4.07SAA677 pKa = 3.51
MM1 pKa = 7.56ALHH4 pKa = 7.13AKK6 pKa = 9.9FLLLVLILLCDD17 pKa = 3.56YY18 pKa = 10.68ARR20 pKa = 11.84SIEE23 pKa = 4.24IVGEE27 pKa = 4.01LRR29 pKa = 11.84NLDD32 pKa = 3.78PGTVKK37 pKa = 10.65GLAARR42 pKa = 11.84GSQLTRR48 pKa = 11.84YY49 pKa = 7.63TNFVQVSRR57 pKa = 11.84PTQGTVRR64 pKa = 11.84EE65 pKa = 4.21DD66 pKa = 3.5HH67 pKa = 6.29YY68 pKa = 11.39VPHH71 pKa = 6.41YY72 pKa = 11.09VDD74 pKa = 4.45IPLDD78 pKa = 3.5TDD80 pKa = 3.65LDD82 pKa = 4.3YY83 pKa = 11.66VVTGTSSLGTDD94 pKa = 4.02FGFRR98 pKa = 11.84VKK100 pKa = 10.65VDD102 pKa = 3.63DD103 pKa = 4.31VNVEE107 pKa = 4.04VEE109 pKa = 4.05NEE111 pKa = 4.38LIYY114 pKa = 9.65TAPFYY119 pKa = 10.71RR120 pKa = 11.84DD121 pKa = 3.06SHH123 pKa = 6.7EE124 pKa = 5.47DD125 pKa = 3.37NLHH128 pKa = 4.98QMVWNWDD135 pKa = 3.58GSSLTHH141 pKa = 6.22GVHH144 pKa = 6.45IPTVCAGLSNVDD156 pKa = 3.46YY157 pKa = 11.53SFFEE161 pKa = 4.41CLRR164 pKa = 11.84TRR166 pKa = 11.84DD167 pKa = 3.48EE168 pKa = 4.3VAQLSQKK175 pKa = 10.54DD176 pKa = 3.82RR177 pKa = 11.84EE178 pKa = 4.39CRR180 pKa = 11.84YY181 pKa = 10.47APLGNEE187 pKa = 3.93YY188 pKa = 10.64LSVGNYY194 pKa = 6.53TVKK197 pKa = 10.63NPRR200 pKa = 11.84GTCLNAEE207 pKa = 4.26SQSWWGRR214 pKa = 11.84PFFTGLYY221 pKa = 6.94KK222 pKa = 10.47TGMPWIRR229 pKa = 11.84AIDD232 pKa = 3.55QWNLADD238 pKa = 5.4CGTGSCTPSGQIRR251 pKa = 11.84VLQRR255 pKa = 11.84KK256 pKa = 7.99WISGVNTAMFFKK268 pKa = 10.31STGKK272 pKa = 10.54AIANYY277 pKa = 8.12TISVSPEE284 pKa = 3.66YY285 pKa = 11.12GCDD288 pKa = 3.6FDD290 pKa = 5.31RR291 pKa = 11.84CEE293 pKa = 4.07LHH295 pKa = 7.43GPGDD299 pKa = 3.67KK300 pKa = 10.0CTLSVAGDD308 pKa = 3.61TMSITGNDD316 pKa = 3.6LEE318 pKa = 4.86TTTRR322 pKa = 11.84MNIMVIAACGDD333 pKa = 3.51YY334 pKa = 10.9SGCQPVAVYY343 pKa = 9.69RR344 pKa = 11.84WPDD347 pKa = 3.43DD348 pKa = 4.35LYY350 pKa = 10.66LTNAFPYY357 pKa = 9.88LKK359 pKa = 9.13WRR361 pKa = 11.84SVPGPVYY368 pKa = 10.77ADD370 pKa = 3.16EE371 pKa = 5.2GKK373 pKa = 8.7TKK375 pKa = 10.36FQADD379 pKa = 3.27AKK381 pKa = 9.95FFPDD385 pKa = 2.81VHH387 pKa = 7.88INEE390 pKa = 4.59EE391 pKa = 4.0FCLDD395 pKa = 3.74LGYY398 pKa = 10.76RR399 pKa = 11.84QDD401 pKa = 3.78EE402 pKa = 5.05GIAQYY407 pKa = 8.32THH409 pKa = 6.75SSNKK413 pKa = 10.55DD414 pKa = 2.59GDD416 pKa = 4.23ANKK419 pKa = 9.45PACRR423 pKa = 11.84KK424 pKa = 8.39KK425 pKa = 10.93APIISQSQRR434 pKa = 11.84EE435 pKa = 4.1FEE437 pKa = 4.13EE438 pKa = 4.62RR439 pKa = 11.84GLSIDD444 pKa = 3.06RR445 pKa = 11.84CGDD448 pKa = 2.75IRR450 pKa = 11.84LEE452 pKa = 4.07YY453 pKa = 10.88VGDD456 pKa = 3.53IDD458 pKa = 5.53MVTLKK463 pKa = 10.67YY464 pKa = 10.94GKK466 pKa = 10.54GYY468 pKa = 10.34DD469 pKa = 3.93GKK471 pKa = 10.66DD472 pKa = 3.31CEE474 pKa = 4.83DD475 pKa = 3.41LKK477 pKa = 11.19KK478 pKa = 11.04VNPHH482 pKa = 6.91PYY484 pKa = 9.66RR485 pKa = 11.84YY486 pKa = 9.24PLSAVKK492 pKa = 9.97AYY494 pKa = 11.25YY495 pKa = 9.83NGCTGGVARR504 pKa = 11.84AYY506 pKa = 10.69FDD508 pKa = 3.32IKK510 pKa = 10.2STEE513 pKa = 4.02IVIDD517 pKa = 4.1PPDD520 pKa = 3.98GVDD523 pKa = 5.49LSVCQIKK530 pKa = 9.38MEE532 pKa = 4.67GYY534 pKa = 9.38YY535 pKa = 11.05GLDD538 pKa = 2.95NGITLTIGGCTVSGPVRR555 pKa = 11.84LQLPQLDD562 pKa = 3.83AYY564 pKa = 10.74FYY566 pKa = 11.15GDD568 pKa = 3.82SKK570 pKa = 10.27TLTTVLGASEE580 pKa = 4.17SRR582 pKa = 11.84ITITMLSYY590 pKa = 11.54YY591 pKa = 10.36DD592 pKa = 3.53VLLIAEE598 pKa = 4.72VFSSTITEE606 pKa = 4.4VPVTALNADD615 pKa = 3.83DD616 pKa = 5.97AYY618 pKa = 9.41TPDD621 pKa = 4.45VIPPPSNDD629 pKa = 2.79GGGFFDD635 pKa = 5.14FLNNMFGNTFSIILIISAVILVGFILVSCCASARR669 pKa = 11.84GGVKK673 pKa = 10.04KK674 pKa = 10.69EE675 pKa = 4.07SAA677 pKa = 3.51
Molecular weight: 74.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIW8|A0A1L3KIW8_9NIDO Putative glycoprotein 1 OS=Beihai Nido-like virus 2 OX=1922351 PE=4 SV=1
MM1 pKa = 7.81PRR3 pKa = 11.84PQPRR7 pKa = 11.84PPSHH11 pKa = 6.59RR12 pKa = 11.84GSRR15 pKa = 11.84EE16 pKa = 3.37VAQAANQTCSPSPTDD31 pKa = 3.19HH32 pKa = 6.99PQPQRR37 pKa = 11.84GIAGVTTEE45 pKa = 4.69AGTTATVAARR55 pKa = 11.84TLITRR60 pKa = 11.84EE61 pKa = 3.92GYY63 pKa = 10.27YY64 pKa = 10.48LLKK67 pKa = 10.97AGDD70 pKa = 4.15LVISRR75 pKa = 11.84QKK77 pKa = 10.98GPPEE81 pKa = 3.87VLLSVPATSRR91 pKa = 11.84VLSGHH96 pKa = 6.25TIGSPP101 pKa = 3.06
MM1 pKa = 7.81PRR3 pKa = 11.84PQPRR7 pKa = 11.84PPSHH11 pKa = 6.59RR12 pKa = 11.84GSRR15 pKa = 11.84EE16 pKa = 3.37VAQAANQTCSPSPTDD31 pKa = 3.19HH32 pKa = 6.99PQPQRR37 pKa = 11.84GIAGVTTEE45 pKa = 4.69AGTTATVAARR55 pKa = 11.84TLITRR60 pKa = 11.84EE61 pKa = 3.92GYY63 pKa = 10.27YY64 pKa = 10.48LLKK67 pKa = 10.97AGDD70 pKa = 4.15LVISRR75 pKa = 11.84QKK77 pKa = 10.98GPPEE81 pKa = 3.87VLLSVPATSRR91 pKa = 11.84VLSGHH96 pKa = 6.25TIGSPP101 pKa = 3.06
Molecular weight: 10.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8003 |
101 |
5487 |
889.2 |
100.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.373 ± 0.345 | 2.512 ± 0.273 |
5.261 ± 0.394 | 4.223 ± 0.274 |
4.286 ± 0.348 | 5.71 ± 0.727 |
3.249 ± 0.314 | 5.948 ± 0.226 |
5.061 ± 0.274 | 9.072 ± 0.371 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.099 ± 0.351 | 4.686 ± 0.233 |
4.798 ± 0.485 | 3.811 ± 0.429 |
4.448 ± 0.69 | 7.06 ± 0.523 |
6.722 ± 0.391 | 7.747 ± 0.586 |
1.025 ± 0.234 | 5.91 ± 0.383 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
