
Aspergillus pseudonomiae
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi;
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
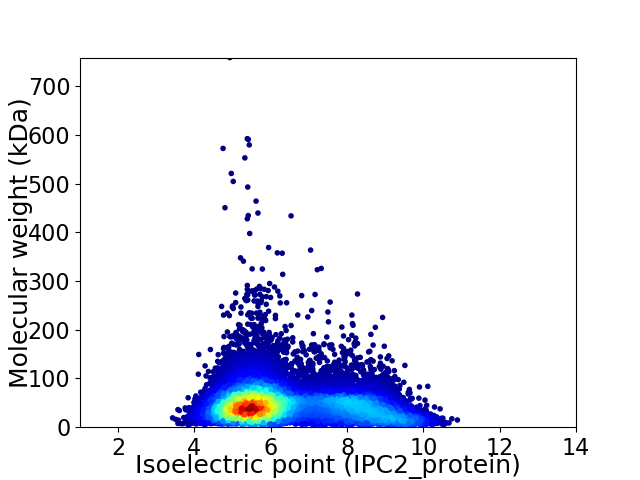
Virtual 2D-PAGE plot for 13384 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5N6IHN6|A0A5N6IHN6_9EURO Uncharacterized protein OS=Aspergillus pseudonomiae OX=1506151 GN=BDV32DRAFT_3048 PE=4 SV=1
MM1 pKa = 7.39YY2 pKa = 10.72LNQLVLAGLALCGSALASPTPAKK25 pKa = 10.44RR26 pKa = 11.84ADD28 pKa = 3.67EE29 pKa = 4.22KK30 pKa = 11.29VGYY33 pKa = 10.41LSVYY37 pKa = 6.69WTTDD41 pKa = 3.14DD42 pKa = 3.55EE43 pKa = 4.89SVYY46 pKa = 10.52FALSDD51 pKa = 3.6NDD53 pKa = 4.0DD54 pKa = 4.0PLGFSTINSGSAVVSPTLGTKK75 pKa = 9.92AVRR78 pKa = 11.84DD79 pKa = 3.6TSIIAGQGDD88 pKa = 3.86DD89 pKa = 4.0AGKK92 pKa = 10.19YY93 pKa = 8.8WIIGTDD99 pKa = 3.63LNIGEE104 pKa = 4.77TTWDD108 pKa = 3.25AAVRR112 pKa = 11.84TGSRR116 pKa = 11.84AIYY119 pKa = 9.3VWEE122 pKa = 4.19STDD125 pKa = 3.71LVNWSEE131 pKa = 4.12NTLVTVEE138 pKa = 4.92DD139 pKa = 3.99STAGMVWAPDD149 pKa = 4.56AIWDD153 pKa = 4.1PEE155 pKa = 3.82QGQYY159 pKa = 9.27FVHH162 pKa = 6.3WASRR166 pKa = 11.84FYY168 pKa = 11.16AADD171 pKa = 3.85DD172 pKa = 4.0TEE174 pKa = 4.5HH175 pKa = 6.64TGDD178 pKa = 3.38ATTGNVLRR186 pKa = 11.84YY187 pKa = 9.93AYY189 pKa = 9.52TSDD192 pKa = 3.08FKK194 pKa = 10.96TFSEE198 pKa = 4.19PQDD201 pKa = 4.06YY202 pKa = 10.67IVGTTDD208 pKa = 4.8VIDD211 pKa = 3.8LCLIQLDD218 pKa = 4.55SNTLLRR224 pKa = 11.84SYY226 pKa = 10.95VNSSSADD233 pKa = 3.47GLPVEE238 pKa = 4.9ISTNGLLGDD247 pKa = 3.87WSLLGSIADD256 pKa = 3.71SAGYY260 pKa = 7.29EE261 pKa = 3.92APYY264 pKa = 10.4FFADD268 pKa = 3.27NAGGGKK274 pKa = 10.58GYY276 pKa = 7.74MTADD280 pKa = 3.72LVGSSPGISGWTSDD294 pKa = 4.14DD295 pKa = 3.65LSSGVLTKK303 pKa = 10.13DD304 pKa = 3.01TSHH307 pKa = 7.41DD308 pKa = 3.67LTFMRR313 pKa = 11.84HH314 pKa = 4.69NSVLAVTQSQYY325 pKa = 11.62DD326 pKa = 3.51ALKK329 pKa = 11.21AMM331 pKa = 5.08
MM1 pKa = 7.39YY2 pKa = 10.72LNQLVLAGLALCGSALASPTPAKK25 pKa = 10.44RR26 pKa = 11.84ADD28 pKa = 3.67EE29 pKa = 4.22KK30 pKa = 11.29VGYY33 pKa = 10.41LSVYY37 pKa = 6.69WTTDD41 pKa = 3.14DD42 pKa = 3.55EE43 pKa = 4.89SVYY46 pKa = 10.52FALSDD51 pKa = 3.6NDD53 pKa = 4.0DD54 pKa = 4.0PLGFSTINSGSAVVSPTLGTKK75 pKa = 9.92AVRR78 pKa = 11.84DD79 pKa = 3.6TSIIAGQGDD88 pKa = 3.86DD89 pKa = 4.0AGKK92 pKa = 10.19YY93 pKa = 8.8WIIGTDD99 pKa = 3.63LNIGEE104 pKa = 4.77TTWDD108 pKa = 3.25AAVRR112 pKa = 11.84TGSRR116 pKa = 11.84AIYY119 pKa = 9.3VWEE122 pKa = 4.19STDD125 pKa = 3.71LVNWSEE131 pKa = 4.12NTLVTVEE138 pKa = 4.92DD139 pKa = 3.99STAGMVWAPDD149 pKa = 4.56AIWDD153 pKa = 4.1PEE155 pKa = 3.82QGQYY159 pKa = 9.27FVHH162 pKa = 6.3WASRR166 pKa = 11.84FYY168 pKa = 11.16AADD171 pKa = 3.85DD172 pKa = 4.0TEE174 pKa = 4.5HH175 pKa = 6.64TGDD178 pKa = 3.38ATTGNVLRR186 pKa = 11.84YY187 pKa = 9.93AYY189 pKa = 9.52TSDD192 pKa = 3.08FKK194 pKa = 10.96TFSEE198 pKa = 4.19PQDD201 pKa = 4.06YY202 pKa = 10.67IVGTTDD208 pKa = 4.8VIDD211 pKa = 3.8LCLIQLDD218 pKa = 4.55SNTLLRR224 pKa = 11.84SYY226 pKa = 10.95VNSSSADD233 pKa = 3.47GLPVEE238 pKa = 4.9ISTNGLLGDD247 pKa = 3.87WSLLGSIADD256 pKa = 3.71SAGYY260 pKa = 7.29EE261 pKa = 3.92APYY264 pKa = 10.4FFADD268 pKa = 3.27NAGGGKK274 pKa = 10.58GYY276 pKa = 7.74MTADD280 pKa = 3.72LVGSSPGISGWTSDD294 pKa = 4.14DD295 pKa = 3.65LSSGVLTKK303 pKa = 10.13DD304 pKa = 3.01TSHH307 pKa = 7.41DD308 pKa = 3.67LTFMRR313 pKa = 11.84HH314 pKa = 4.69NSVLAVTQSQYY325 pKa = 11.62DD326 pKa = 3.51ALKK329 pKa = 11.21AMM331 pKa = 5.08
Molecular weight: 35.51 kDa
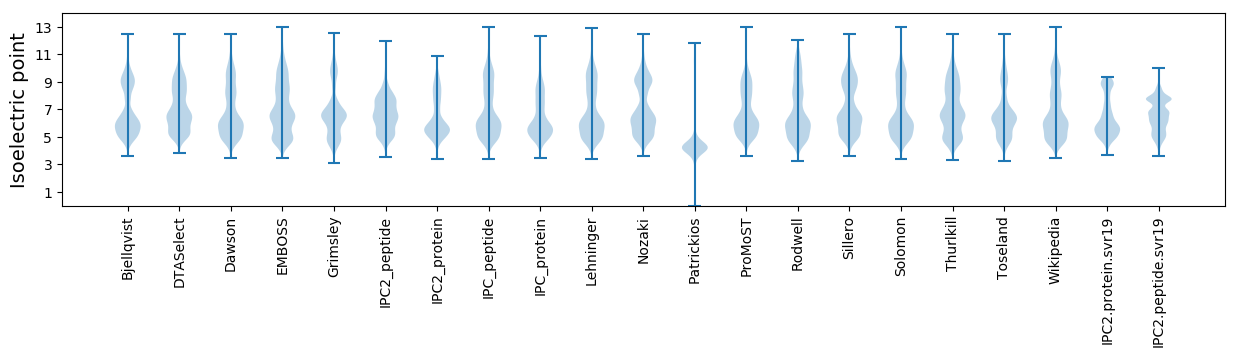
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5N6HXM4|A0A5N6HXM4_9EURO Uncharacterized protein OS=Aspergillus pseudonomiae OX=1506151 GN=BDV32DRAFT_150549 PE=4 SV=1
MM1 pKa = 7.96DD2 pKa = 6.01SIGSTGRR9 pKa = 11.84GRR11 pKa = 11.84NKK13 pKa = 9.9IQLLKK18 pKa = 9.35PSRR21 pKa = 11.84MRR23 pKa = 11.84SRR25 pKa = 11.84TLVHH29 pKa = 5.19VTFSGVLLRR38 pKa = 11.84LLTGLLLVYY47 pKa = 10.16RR48 pKa = 11.84HH49 pKa = 6.58SILRR53 pKa = 11.84AYY55 pKa = 9.97VIGYY59 pKa = 9.58LSSTTPRR66 pKa = 11.84VIASLRR72 pKa = 11.84QIWRR76 pKa = 11.84NDD78 pKa = 2.78LDD80 pKa = 3.91YY81 pKa = 10.36QQKK84 pKa = 10.04RR85 pKa = 11.84RR86 pKa = 11.84VLVRR90 pKa = 11.84ALTSAIRR97 pKa = 11.84LDD99 pKa = 3.91SFPTFCASLVGGSTVFPLVIFRR121 pKa = 11.84LAEE124 pKa = 4.06LLSQKK129 pKa = 10.29VDD131 pKa = 4.51FKK133 pKa = 11.2CSRR136 pKa = 11.84LEE138 pKa = 3.66SARR141 pKa = 11.84FLRR144 pKa = 11.84LARR147 pKa = 11.84LICTFVSAWLSFEE160 pKa = 4.12LLNRR164 pKa = 11.84KK165 pKa = 8.93PIRR168 pKa = 11.84LQGIEE173 pKa = 4.28GTLADD178 pKa = 4.35NGSDD182 pKa = 3.26EE183 pKa = 5.23AAGKK187 pKa = 8.45QQGCQNTIHH196 pKa = 6.42NRR198 pKa = 11.84PQFAGRR204 pKa = 11.84TMDD207 pKa = 3.33LTLFSFTRR215 pKa = 11.84AVDD218 pKa = 4.12LVACIIWARR227 pKa = 11.84WRR229 pKa = 11.84RR230 pKa = 11.84WRR232 pKa = 11.84SARR235 pKa = 11.84GRR237 pKa = 11.84WSRR240 pKa = 11.84AEE242 pKa = 3.86SLAPKK247 pKa = 9.11LTDD250 pKa = 2.9SGVFAASAAIVMWAWFYY267 pKa = 11.4LPEE270 pKa = 4.93RR271 pKa = 11.84LPKK274 pKa = 10.42SYY276 pKa = 10.54GKK278 pKa = 9.77WIGEE282 pKa = 4.12VAKK285 pKa = 10.63VDD287 pKa = 3.53SRR289 pKa = 11.84LIEE292 pKa = 3.67ALRR295 pKa = 11.84RR296 pKa = 11.84VRR298 pKa = 11.84RR299 pKa = 11.84GVFVYY304 pKa = 10.28GKK306 pKa = 8.77DD307 pKa = 3.14TGQAPLLQSMCKK319 pKa = 10.25DD320 pKa = 3.61YY321 pKa = 10.93GWPIEE326 pKa = 4.2WGDD329 pKa = 3.57PAKK332 pKa = 10.04TIPIPCEE339 pKa = 3.88MVHH342 pKa = 6.01MSCGPNCEE350 pKa = 3.69KK351 pKa = 10.63HH352 pKa = 5.5AVSRR356 pKa = 11.84FVRR359 pKa = 11.84TFGFACATYY368 pKa = 10.22IPLQILFRR376 pKa = 11.84LKK378 pKa = 10.31RR379 pKa = 11.84LKK381 pKa = 10.66SVLSLRR387 pKa = 11.84RR388 pKa = 11.84AVSDD392 pKa = 3.56ALRR395 pKa = 11.84SSVFLASFVSIFYY408 pKa = 10.75YY409 pKa = 9.68SVCLARR415 pKa = 11.84TRR417 pKa = 11.84IGPKK421 pKa = 9.43VFPRR425 pKa = 11.84DD426 pKa = 3.24VVTPMMWDD434 pKa = 2.97SGLCVGAGCLMCGWSILVEE453 pKa = 4.44SPSKK457 pKa = 10.38RR458 pKa = 11.84QEE460 pKa = 3.62LALFVAPRR468 pKa = 11.84AAATVLPRR476 pKa = 11.84FYY478 pKa = 10.89DD479 pKa = 2.98KK480 pKa = 10.71QYY482 pKa = 10.8QYY484 pKa = 11.29RR485 pKa = 11.84EE486 pKa = 4.17RR487 pKa = 11.84ITFAVSAALLLTCLQEE503 pKa = 4.15RR504 pKa = 11.84PGLVRR509 pKa = 11.84GVFGRR514 pKa = 11.84IATSVLKK521 pKa = 10.85
MM1 pKa = 7.96DD2 pKa = 6.01SIGSTGRR9 pKa = 11.84GRR11 pKa = 11.84NKK13 pKa = 9.9IQLLKK18 pKa = 9.35PSRR21 pKa = 11.84MRR23 pKa = 11.84SRR25 pKa = 11.84TLVHH29 pKa = 5.19VTFSGVLLRR38 pKa = 11.84LLTGLLLVYY47 pKa = 10.16RR48 pKa = 11.84HH49 pKa = 6.58SILRR53 pKa = 11.84AYY55 pKa = 9.97VIGYY59 pKa = 9.58LSSTTPRR66 pKa = 11.84VIASLRR72 pKa = 11.84QIWRR76 pKa = 11.84NDD78 pKa = 2.78LDD80 pKa = 3.91YY81 pKa = 10.36QQKK84 pKa = 10.04RR85 pKa = 11.84RR86 pKa = 11.84VLVRR90 pKa = 11.84ALTSAIRR97 pKa = 11.84LDD99 pKa = 3.91SFPTFCASLVGGSTVFPLVIFRR121 pKa = 11.84LAEE124 pKa = 4.06LLSQKK129 pKa = 10.29VDD131 pKa = 4.51FKK133 pKa = 11.2CSRR136 pKa = 11.84LEE138 pKa = 3.66SARR141 pKa = 11.84FLRR144 pKa = 11.84LARR147 pKa = 11.84LICTFVSAWLSFEE160 pKa = 4.12LLNRR164 pKa = 11.84KK165 pKa = 8.93PIRR168 pKa = 11.84LQGIEE173 pKa = 4.28GTLADD178 pKa = 4.35NGSDD182 pKa = 3.26EE183 pKa = 5.23AAGKK187 pKa = 8.45QQGCQNTIHH196 pKa = 6.42NRR198 pKa = 11.84PQFAGRR204 pKa = 11.84TMDD207 pKa = 3.33LTLFSFTRR215 pKa = 11.84AVDD218 pKa = 4.12LVACIIWARR227 pKa = 11.84WRR229 pKa = 11.84RR230 pKa = 11.84WRR232 pKa = 11.84SARR235 pKa = 11.84GRR237 pKa = 11.84WSRR240 pKa = 11.84AEE242 pKa = 3.86SLAPKK247 pKa = 9.11LTDD250 pKa = 2.9SGVFAASAAIVMWAWFYY267 pKa = 11.4LPEE270 pKa = 4.93RR271 pKa = 11.84LPKK274 pKa = 10.42SYY276 pKa = 10.54GKK278 pKa = 9.77WIGEE282 pKa = 4.12VAKK285 pKa = 10.63VDD287 pKa = 3.53SRR289 pKa = 11.84LIEE292 pKa = 3.67ALRR295 pKa = 11.84RR296 pKa = 11.84VRR298 pKa = 11.84RR299 pKa = 11.84GVFVYY304 pKa = 10.28GKK306 pKa = 8.77DD307 pKa = 3.14TGQAPLLQSMCKK319 pKa = 10.25DD320 pKa = 3.61YY321 pKa = 10.93GWPIEE326 pKa = 4.2WGDD329 pKa = 3.57PAKK332 pKa = 10.04TIPIPCEE339 pKa = 3.88MVHH342 pKa = 6.01MSCGPNCEE350 pKa = 3.69KK351 pKa = 10.63HH352 pKa = 5.5AVSRR356 pKa = 11.84FVRR359 pKa = 11.84TFGFACATYY368 pKa = 10.22IPLQILFRR376 pKa = 11.84LKK378 pKa = 10.31RR379 pKa = 11.84LKK381 pKa = 10.66SVLSLRR387 pKa = 11.84RR388 pKa = 11.84AVSDD392 pKa = 3.56ALRR395 pKa = 11.84SSVFLASFVSIFYY408 pKa = 10.75YY409 pKa = 9.68SVCLARR415 pKa = 11.84TRR417 pKa = 11.84IGPKK421 pKa = 9.43VFPRR425 pKa = 11.84DD426 pKa = 3.24VVTPMMWDD434 pKa = 2.97SGLCVGAGCLMCGWSILVEE453 pKa = 4.44SPSKK457 pKa = 10.38RR458 pKa = 11.84QEE460 pKa = 3.62LALFVAPRR468 pKa = 11.84AAATVLPRR476 pKa = 11.84FYY478 pKa = 10.89DD479 pKa = 2.98KK480 pKa = 10.71QYY482 pKa = 10.8QYY484 pKa = 11.29RR485 pKa = 11.84EE486 pKa = 4.17RR487 pKa = 11.84ITFAVSAALLLTCLQEE503 pKa = 4.15RR504 pKa = 11.84PGLVRR509 pKa = 11.84GVFGRR514 pKa = 11.84IATSVLKK521 pKa = 10.85
Molecular weight: 58.82 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6142267 |
49 |
6889 |
458.9 |
50.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.247 ± 0.017 | 1.374 ± 0.009 |
5.563 ± 0.015 | 5.999 ± 0.021 |
3.86 ± 0.013 | 6.789 ± 0.015 |
2.469 ± 0.009 | 5.146 ± 0.017 |
4.539 ± 0.016 | 9.255 ± 0.023 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.181 ± 0.008 | 3.678 ± 0.008 |
5.888 ± 0.022 | 4.023 ± 0.015 |
6.031 ± 0.018 | 8.251 ± 0.025 |
5.934 ± 0.013 | 6.273 ± 0.016 |
1.536 ± 0.008 | 2.962 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
