
Hybrid snakehead virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; unclassified Rhabdoviridae; Siniperca chuatsi rhabdovirus
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
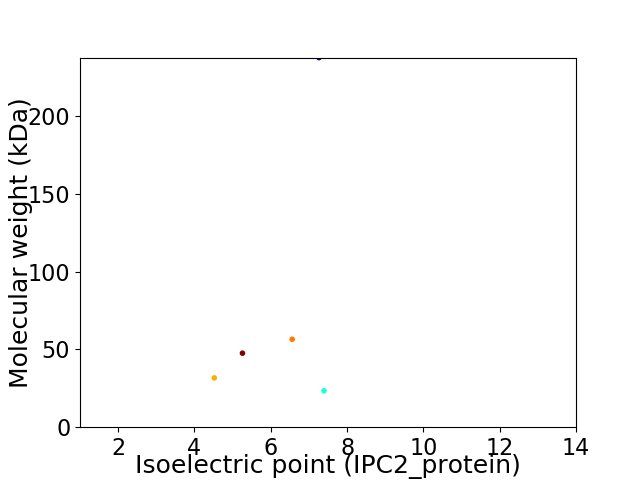
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M9T5K3|M9T5K3_9RHAB GDP polyribonucleotidyltransferase OS=Hybrid snakehead virus OX=1316453 GN=L PE=4 SV=1
MM1 pKa = 7.82AKK3 pKa = 9.93PVFQISYY10 pKa = 10.12DD11 pKa = 3.89LEE13 pKa = 4.69KK14 pKa = 10.78LATNFEE20 pKa = 4.36EE21 pKa = 5.47LNLAEE26 pKa = 4.67LKK28 pKa = 10.94RR29 pKa = 11.84NGEE32 pKa = 4.29DD33 pKa = 3.66PDD35 pKa = 5.66DD36 pKa = 4.41PLVWKK41 pKa = 9.22PSPDD45 pKa = 3.69LLPYY49 pKa = 9.97AASPEE54 pKa = 4.07EE55 pKa = 3.98EE56 pKa = 4.61EE57 pKa = 4.53YY58 pKa = 11.03EE59 pKa = 4.07QGEE62 pKa = 4.26QEE64 pKa = 4.22MEE66 pKa = 4.26CTDD69 pKa = 3.93EE70 pKa = 3.99YY71 pKa = 11.37TPADD75 pKa = 3.9SSWTAEE81 pKa = 4.05VSRR84 pKa = 11.84PPILPGYY91 pKa = 5.58TTQSRR96 pKa = 11.84WFGPGVTAEE105 pKa = 4.26TKK107 pKa = 10.38PIILEE112 pKa = 3.92EE113 pKa = 4.1LASVLQQIGIKK124 pKa = 9.88LISAAPSADD133 pKa = 2.91ANYY136 pKa = 10.66YY137 pKa = 10.38LLYY140 pKa = 10.23PNIHH144 pKa = 6.53MSLMAPGVLEE154 pKa = 4.02SGRR157 pKa = 11.84PLTPYY162 pKa = 7.8TAPAEE167 pKa = 4.31APKK170 pKa = 10.36PPAAPAPDD178 pKa = 4.2PPKK181 pKa = 10.15EE182 pKa = 4.29SPPAPRR188 pKa = 11.84QPVPALAEE196 pKa = 3.88QSPTGLILNDD206 pKa = 3.65DD207 pKa = 4.24DD208 pKa = 4.13VRR210 pKa = 11.84AVLWASGVRR219 pKa = 11.84VVEE222 pKa = 4.14KK223 pKa = 9.66KK224 pKa = 8.77TNKK227 pKa = 8.67TAILKK232 pKa = 9.4PCALGWMIDD241 pKa = 3.34DD242 pKa = 4.58WLQVDD247 pKa = 4.27IGAEE251 pKa = 3.88TDD253 pKa = 3.27VAEE256 pKa = 4.56IFKK259 pKa = 11.04KK260 pKa = 10.1MLAKK264 pKa = 9.79TPSRR268 pKa = 11.84KK269 pKa = 10.17LMMYY273 pKa = 9.94KK274 pKa = 10.33YY275 pKa = 10.13IPLEE279 pKa = 4.01VVQQMVLFF287 pKa = 4.7
MM1 pKa = 7.82AKK3 pKa = 9.93PVFQISYY10 pKa = 10.12DD11 pKa = 3.89LEE13 pKa = 4.69KK14 pKa = 10.78LATNFEE20 pKa = 4.36EE21 pKa = 5.47LNLAEE26 pKa = 4.67LKK28 pKa = 10.94RR29 pKa = 11.84NGEE32 pKa = 4.29DD33 pKa = 3.66PDD35 pKa = 5.66DD36 pKa = 4.41PLVWKK41 pKa = 9.22PSPDD45 pKa = 3.69LLPYY49 pKa = 9.97AASPEE54 pKa = 4.07EE55 pKa = 3.98EE56 pKa = 4.61EE57 pKa = 4.53YY58 pKa = 11.03EE59 pKa = 4.07QGEE62 pKa = 4.26QEE64 pKa = 4.22MEE66 pKa = 4.26CTDD69 pKa = 3.93EE70 pKa = 3.99YY71 pKa = 11.37TPADD75 pKa = 3.9SSWTAEE81 pKa = 4.05VSRR84 pKa = 11.84PPILPGYY91 pKa = 5.58TTQSRR96 pKa = 11.84WFGPGVTAEE105 pKa = 4.26TKK107 pKa = 10.38PIILEE112 pKa = 3.92EE113 pKa = 4.1LASVLQQIGIKK124 pKa = 9.88LISAAPSADD133 pKa = 2.91ANYY136 pKa = 10.66YY137 pKa = 10.38LLYY140 pKa = 10.23PNIHH144 pKa = 6.53MSLMAPGVLEE154 pKa = 4.02SGRR157 pKa = 11.84PLTPYY162 pKa = 7.8TAPAEE167 pKa = 4.31APKK170 pKa = 10.36PPAAPAPDD178 pKa = 4.2PPKK181 pKa = 10.15EE182 pKa = 4.29SPPAPRR188 pKa = 11.84QPVPALAEE196 pKa = 3.88QSPTGLILNDD206 pKa = 3.65DD207 pKa = 4.24DD208 pKa = 4.13VRR210 pKa = 11.84AVLWASGVRR219 pKa = 11.84VVEE222 pKa = 4.14KK223 pKa = 9.66KK224 pKa = 8.77TNKK227 pKa = 8.67TAILKK232 pKa = 9.4PCALGWMIDD241 pKa = 3.34DD242 pKa = 4.58WLQVDD247 pKa = 4.27IGAEE251 pKa = 3.88TDD253 pKa = 3.27VAEE256 pKa = 4.56IFKK259 pKa = 11.04KK260 pKa = 10.1MLAKK264 pKa = 9.79TPSRR268 pKa = 11.84KK269 pKa = 10.17LMMYY273 pKa = 9.94KK274 pKa = 10.33YY275 pKa = 10.13IPLEE279 pKa = 4.01VVQQMVLFF287 pKa = 4.7
Molecular weight: 31.7 kDa
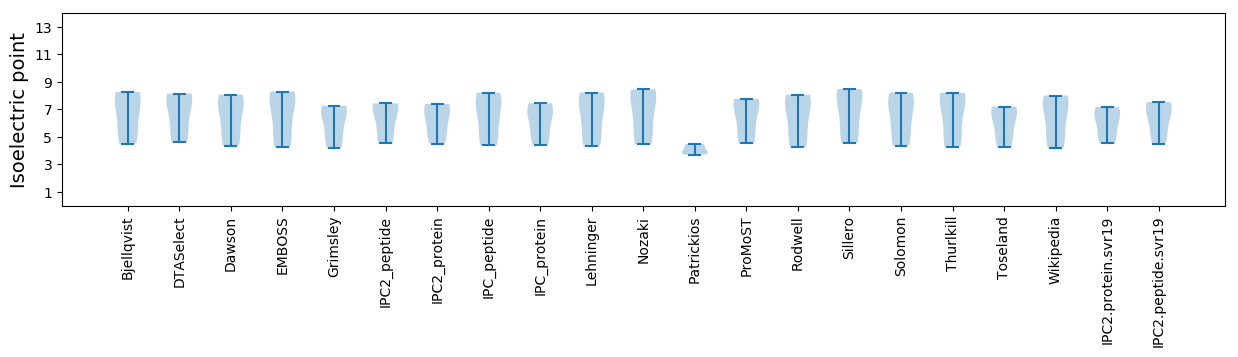
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M9T1J2|M9T1J2_9RHAB Nucleocapsid protein OS=Hybrid snakehead virus OX=1316453 GN=N PE=4 SV=1
MM1 pKa = 7.79PLFKK5 pKa = 10.62KK6 pKa = 10.49SNKK9 pKa = 9.04KK10 pKa = 10.33SAVKK14 pKa = 9.84PYY16 pKa = 9.74QAPPPYY22 pKa = 9.98SATALTPSAPMDD34 pKa = 4.57LPDD37 pKa = 3.41NDD39 pKa = 3.91YY40 pKa = 10.8RR41 pKa = 11.84IKK43 pKa = 10.45TMMVEE48 pKa = 4.65LDD50 pKa = 3.66FKK52 pKa = 9.88ITSSIEE58 pKa = 3.58LKK60 pKa = 10.17TIGKK64 pKa = 9.49IYY66 pKa = 10.49QIAQYY71 pKa = 10.16MLDD74 pKa = 4.28EE75 pKa = 4.24YY76 pKa = 10.57TGPIRR81 pKa = 11.84SKK83 pKa = 9.84PLYY86 pKa = 9.08MGLFLASCHH95 pKa = 5.37NAINPSMVHH104 pKa = 5.99GKK106 pKa = 6.85WHH108 pKa = 6.18YY109 pKa = 9.47EE110 pKa = 3.52IQFRR114 pKa = 11.84GPIGFNLANNTPLDD128 pKa = 3.9WVCNPVAISYY138 pKa = 7.69EE139 pKa = 4.34CNTPEE144 pKa = 4.32RR145 pKa = 11.84SLVSYY150 pKa = 7.8TCNMRR155 pKa = 11.84PTKK158 pKa = 8.7MTGSSFEE165 pKa = 4.19KK166 pKa = 9.51MFHH169 pKa = 6.02GVLVHH174 pKa = 6.53PSAASVFGIFQMAEE188 pKa = 3.7AEE190 pKa = 4.35VRR192 pKa = 11.84GEE194 pKa = 4.38DD195 pKa = 3.58IVFILKK201 pKa = 10.56DD202 pKa = 3.92PGHH205 pKa = 6.18SWHH208 pKa = 6.92
MM1 pKa = 7.79PLFKK5 pKa = 10.62KK6 pKa = 10.49SNKK9 pKa = 9.04KK10 pKa = 10.33SAVKK14 pKa = 9.84PYY16 pKa = 9.74QAPPPYY22 pKa = 9.98SATALTPSAPMDD34 pKa = 4.57LPDD37 pKa = 3.41NDD39 pKa = 3.91YY40 pKa = 10.8RR41 pKa = 11.84IKK43 pKa = 10.45TMMVEE48 pKa = 4.65LDD50 pKa = 3.66FKK52 pKa = 9.88ITSSIEE58 pKa = 3.58LKK60 pKa = 10.17TIGKK64 pKa = 9.49IYY66 pKa = 10.49QIAQYY71 pKa = 10.16MLDD74 pKa = 4.28EE75 pKa = 4.24YY76 pKa = 10.57TGPIRR81 pKa = 11.84SKK83 pKa = 9.84PLYY86 pKa = 9.08MGLFLASCHH95 pKa = 5.37NAINPSMVHH104 pKa = 5.99GKK106 pKa = 6.85WHH108 pKa = 6.18YY109 pKa = 9.47EE110 pKa = 3.52IQFRR114 pKa = 11.84GPIGFNLANNTPLDD128 pKa = 3.9WVCNPVAISYY138 pKa = 7.69EE139 pKa = 4.34CNTPEE144 pKa = 4.32RR145 pKa = 11.84SLVSYY150 pKa = 7.8TCNMRR155 pKa = 11.84PTKK158 pKa = 8.7MTGSSFEE165 pKa = 4.19KK166 pKa = 9.51MFHH169 pKa = 6.02GVLVHH174 pKa = 6.53PSAASVFGIFQMAEE188 pKa = 3.7AEE190 pKa = 4.35VRR192 pKa = 11.84GEE194 pKa = 4.38DD195 pKa = 3.58IVFILKK201 pKa = 10.56DD202 pKa = 3.92PGHH205 pKa = 6.18SWHH208 pKa = 6.92
Molecular weight: 23.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3510 |
208 |
2078 |
702.0 |
79.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.496 ± 1.421 | 1.51 ± 0.299 |
5.271 ± 0.359 | 6.581 ± 0.419 |
3.932 ± 0.382 | 5.84 ± 0.408 |
2.536 ± 0.496 | 5.954 ± 0.162 |
6.581 ± 0.517 | 10.427 ± 0.715 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.162 ± 0.322 | 4.501 ± 0.288 |
4.986 ± 1.211 | 3.675 ± 0.199 |
4.786 ± 0.541 | 7.607 ± 0.679 |
5.755 ± 0.309 | 5.242 ± 0.495 |
1.852 ± 0.12 | 3.305 ± 0.348 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
