
Maize rough dwarf virus (MRDV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Fijivirus
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
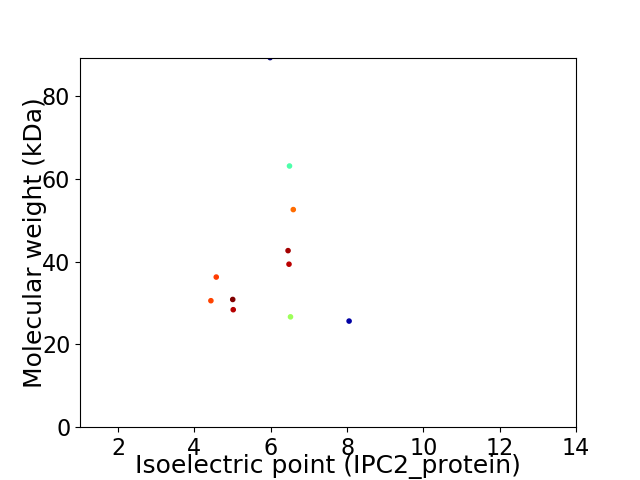
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

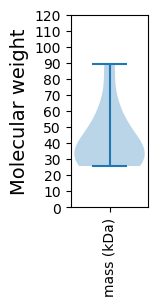
Protein with the lowest isoelectric point:
>tr|A0A077X598|A0A077X598_MRDV Uncharacterized protein (Fragment) OS=Maize rough dwarf virus OX=10989 GN=P5-1 PE=4 SV=1
SS1 pKa = 7.07YY2 pKa = 11.01EE3 pKa = 4.43DD4 pKa = 3.91FLTNVKK10 pKa = 10.15LVEE13 pKa = 3.88FAKK16 pKa = 10.22RR17 pKa = 11.84YY18 pKa = 9.88SSEE21 pKa = 3.73TDD23 pKa = 3.23DD24 pKa = 3.57LHH26 pKa = 7.22RR27 pKa = 11.84ATIEE31 pKa = 3.76LARR34 pKa = 11.84MSEE37 pKa = 4.2EE38 pKa = 3.82NGRR41 pKa = 11.84LLVTLKK47 pKa = 8.98QTQEE51 pKa = 3.9RR52 pKa = 11.84IANFEE57 pKa = 4.24AEE59 pKa = 4.56VMDD62 pKa = 5.04LEE64 pKa = 4.3KK65 pKa = 10.97QRR67 pKa = 11.84DD68 pKa = 4.06CFDD71 pKa = 3.84LQLTEE76 pKa = 5.97LNDD79 pKa = 4.15AYY81 pKa = 11.13CSLIKK86 pKa = 10.64EE87 pKa = 3.95KK88 pKa = 11.32DD89 pKa = 3.43EE90 pKa = 4.12IKK92 pKa = 10.15TEE94 pKa = 3.85NEE96 pKa = 3.7MLKK99 pKa = 10.29EE100 pKa = 3.84QLKK103 pKa = 9.18EE104 pKa = 3.87ANNVVVEE111 pKa = 4.51RR112 pKa = 11.84TARR115 pKa = 11.84LNEE118 pKa = 4.86AIDD121 pKa = 3.41EE122 pKa = 4.34LEE124 pKa = 4.09NVKK127 pKa = 8.76EE128 pKa = 4.15TGNAVTEE135 pKa = 4.17EE136 pKa = 4.1SDD138 pKa = 3.87EE139 pKa = 4.33VEE141 pKa = 4.25HH142 pKa = 6.73EE143 pKa = 4.16SDD145 pKa = 3.31GFRR148 pKa = 11.84LFGEE152 pKa = 5.13CIDD155 pKa = 3.72KK156 pKa = 10.78VRR158 pKa = 11.84ALKK161 pKa = 10.24EE162 pKa = 4.25KK163 pKa = 10.63YY164 pKa = 7.54PHH166 pKa = 7.04LYY168 pKa = 10.56SGVDD172 pKa = 3.31ANFGDD177 pKa = 3.81ITSLLKK183 pKa = 10.29MIMIKK188 pKa = 9.6IDD190 pKa = 4.22SIMSLCDD197 pKa = 3.25ADD199 pKa = 3.92VTVSEE204 pKa = 4.62NQWNAYY210 pKa = 7.43GLCSSVSTSTDD221 pKa = 3.29DD222 pKa = 3.93NSFPMLSVRR231 pKa = 11.84QVGLMNVQIANFTNVIYY248 pKa = 10.58DD249 pKa = 3.4AFFYY253 pKa = 11.42NMFIKK258 pKa = 10.04FLWQLSSEE266 pKa = 4.32
SS1 pKa = 7.07YY2 pKa = 11.01EE3 pKa = 4.43DD4 pKa = 3.91FLTNVKK10 pKa = 10.15LVEE13 pKa = 3.88FAKK16 pKa = 10.22RR17 pKa = 11.84YY18 pKa = 9.88SSEE21 pKa = 3.73TDD23 pKa = 3.23DD24 pKa = 3.57LHH26 pKa = 7.22RR27 pKa = 11.84ATIEE31 pKa = 3.76LARR34 pKa = 11.84MSEE37 pKa = 4.2EE38 pKa = 3.82NGRR41 pKa = 11.84LLVTLKK47 pKa = 8.98QTQEE51 pKa = 3.9RR52 pKa = 11.84IANFEE57 pKa = 4.24AEE59 pKa = 4.56VMDD62 pKa = 5.04LEE64 pKa = 4.3KK65 pKa = 10.97QRR67 pKa = 11.84DD68 pKa = 4.06CFDD71 pKa = 3.84LQLTEE76 pKa = 5.97LNDD79 pKa = 4.15AYY81 pKa = 11.13CSLIKK86 pKa = 10.64EE87 pKa = 3.95KK88 pKa = 11.32DD89 pKa = 3.43EE90 pKa = 4.12IKK92 pKa = 10.15TEE94 pKa = 3.85NEE96 pKa = 3.7MLKK99 pKa = 10.29EE100 pKa = 3.84QLKK103 pKa = 9.18EE104 pKa = 3.87ANNVVVEE111 pKa = 4.51RR112 pKa = 11.84TARR115 pKa = 11.84LNEE118 pKa = 4.86AIDD121 pKa = 3.41EE122 pKa = 4.34LEE124 pKa = 4.09NVKK127 pKa = 8.76EE128 pKa = 4.15TGNAVTEE135 pKa = 4.17EE136 pKa = 4.1SDD138 pKa = 3.87EE139 pKa = 4.33VEE141 pKa = 4.25HH142 pKa = 6.73EE143 pKa = 4.16SDD145 pKa = 3.31GFRR148 pKa = 11.84LFGEE152 pKa = 5.13CIDD155 pKa = 3.72KK156 pKa = 10.78VRR158 pKa = 11.84ALKK161 pKa = 10.24EE162 pKa = 4.25KK163 pKa = 10.63YY164 pKa = 7.54PHH166 pKa = 7.04LYY168 pKa = 10.56SGVDD172 pKa = 3.31ANFGDD177 pKa = 3.81ITSLLKK183 pKa = 10.29MIMIKK188 pKa = 9.6IDD190 pKa = 4.22SIMSLCDD197 pKa = 3.25ADD199 pKa = 3.92VTVSEE204 pKa = 4.62NQWNAYY210 pKa = 7.43GLCSSVSTSTDD221 pKa = 3.29DD222 pKa = 3.93NSFPMLSVRR231 pKa = 11.84QVGLMNVQIANFTNVIYY248 pKa = 10.58DD249 pKa = 3.4AFFYY253 pKa = 11.42NMFIKK258 pKa = 10.04FLWQLSSEE266 pKa = 4.32
Molecular weight: 30.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A077X4P5|A0A077X4P5_MRDV Isoform of A0A077X598 Protein P5-1 (Fragment) OS=Maize rough dwarf virus OX=10989 GN=P5-1 PE=4 SV=1
AA1 pKa = 7.86LLRR4 pKa = 11.84SSEE7 pKa = 4.1AFPVNAYY14 pKa = 9.3KK15 pKa = 10.91VSVGWNGNSFAPEE28 pKa = 3.63ITDD31 pKa = 3.19PTMIGDD37 pKa = 3.58VSLIKK42 pKa = 10.35NVSYY46 pKa = 11.24VGVHH50 pKa = 6.25SSDD53 pKa = 3.24NYY55 pKa = 10.58KK56 pKa = 10.28DD57 pKa = 3.51RR58 pKa = 11.84KK59 pKa = 8.82MFSRR63 pKa = 11.84LKK65 pKa = 9.84TFLEE69 pKa = 4.18IAANCNLVLKK79 pKa = 10.32GCRR82 pKa = 11.84TKK84 pKa = 10.61PLSIEE89 pKa = 3.98YY90 pKa = 10.6DD91 pKa = 3.12NTRR94 pKa = 11.84ISKK97 pKa = 10.25KK98 pKa = 10.38LAGKK102 pKa = 9.95KK103 pKa = 9.41ILYY106 pKa = 9.45NDD108 pKa = 3.6RR109 pKa = 11.84AHH111 pKa = 7.35IIFNGIYY118 pKa = 9.91HH119 pKa = 7.29AVTPFMNKK127 pKa = 9.16VNDD130 pKa = 3.62MLMDD134 pKa = 4.87GNTEE138 pKa = 3.56IDD140 pKa = 3.09FDD142 pKa = 4.48KK143 pKa = 10.48IYY145 pKa = 11.05KK146 pKa = 10.01FGRR149 pKa = 11.84DD150 pKa = 2.97AEE152 pKa = 4.35KK153 pKa = 10.64RR154 pKa = 11.84INTYY158 pKa = 11.66DD159 pKa = 3.75MICRR163 pKa = 11.84GYY165 pKa = 11.34GNMDD169 pKa = 3.2QFSSYY174 pKa = 10.64LVNLSTQRR182 pKa = 11.84ICDD185 pKa = 3.32NRR187 pKa = 11.84MTANHH192 pKa = 6.62YY193 pKa = 7.27KK194 pKa = 9.72TLMAANYY201 pKa = 6.8PNRR204 pKa = 11.84VSMHH208 pKa = 5.86VFDD211 pKa = 5.39TMIDD215 pKa = 3.58DD216 pKa = 5.03LLFVYY221 pKa = 10.58CPTT224 pKa = 3.95
AA1 pKa = 7.86LLRR4 pKa = 11.84SSEE7 pKa = 4.1AFPVNAYY14 pKa = 9.3KK15 pKa = 10.91VSVGWNGNSFAPEE28 pKa = 3.63ITDD31 pKa = 3.19PTMIGDD37 pKa = 3.58VSLIKK42 pKa = 10.35NVSYY46 pKa = 11.24VGVHH50 pKa = 6.25SSDD53 pKa = 3.24NYY55 pKa = 10.58KK56 pKa = 10.28DD57 pKa = 3.51RR58 pKa = 11.84KK59 pKa = 8.82MFSRR63 pKa = 11.84LKK65 pKa = 9.84TFLEE69 pKa = 4.18IAANCNLVLKK79 pKa = 10.32GCRR82 pKa = 11.84TKK84 pKa = 10.61PLSIEE89 pKa = 3.98YY90 pKa = 10.6DD91 pKa = 3.12NTRR94 pKa = 11.84ISKK97 pKa = 10.25KK98 pKa = 10.38LAGKK102 pKa = 9.95KK103 pKa = 9.41ILYY106 pKa = 9.45NDD108 pKa = 3.6RR109 pKa = 11.84AHH111 pKa = 7.35IIFNGIYY118 pKa = 9.91HH119 pKa = 7.29AVTPFMNKK127 pKa = 9.16VNDD130 pKa = 3.62MLMDD134 pKa = 4.87GNTEE138 pKa = 3.56IDD140 pKa = 3.09FDD142 pKa = 4.48KK143 pKa = 10.48IYY145 pKa = 11.05KK146 pKa = 10.01FGRR149 pKa = 11.84DD150 pKa = 2.97AEE152 pKa = 4.35KK153 pKa = 10.64RR154 pKa = 11.84INTYY158 pKa = 11.66DD159 pKa = 3.75MICRR163 pKa = 11.84GYY165 pKa = 11.34GNMDD169 pKa = 3.2QFSSYY174 pKa = 10.64LVNLSTQRR182 pKa = 11.84ICDD185 pKa = 3.32NRR187 pKa = 11.84MTANHH192 pKa = 6.62YY193 pKa = 7.27KK194 pKa = 9.72TLMAANYY201 pKa = 6.8PNRR204 pKa = 11.84VSMHH208 pKa = 5.86VFDD211 pKa = 5.39TMIDD215 pKa = 3.58DD216 pKa = 5.03LLFVYY221 pKa = 10.58CPTT224 pKa = 3.95
Molecular weight: 25.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4054 |
224 |
778 |
368.5 |
42.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.169 ± 0.368 | 1.727 ± 0.309 |
6.438 ± 0.302 | 5.945 ± 0.673 |
5.895 ± 0.406 | 3.823 ± 0.336 |
2.96 ± 0.323 | 6.931 ± 0.522 |
6.438 ± 0.294 | 10.927 ± 0.455 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.516 ± 0.316 | 6.29 ± 0.266 |
3.256 ± 0.304 | 2.664 ± 0.209 |
4.243 ± 0.254 | 8.781 ± 0.776 |
5.673 ± 0.509 | 6.438 ± 0.444 |
0.641 ± 0.165 | 4.243 ± 0.334 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
