
Brevibacterium yomogidense
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Brevibacteriaceae; Brevibacterium
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
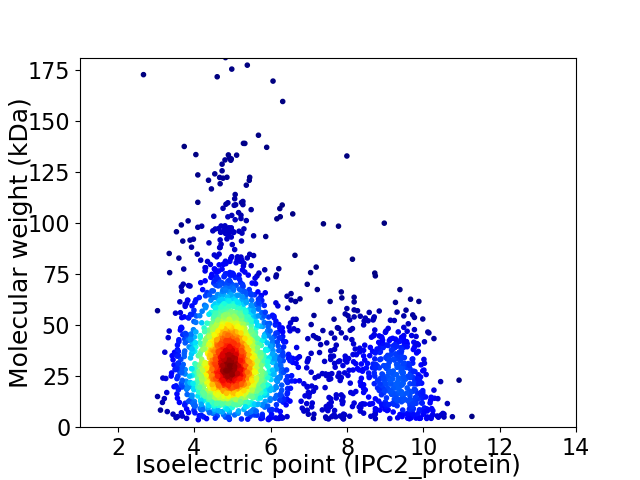
Virtual 2D-PAGE plot for 2868 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1X6XIM2|A0A1X6XIM2_9MICO Glycolate dehydrogenase subunit GlcD OS=Brevibacterium yomogidense OX=946573 GN=FM105_10355 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 9.74PSRR5 pKa = 11.84ISPRR9 pKa = 11.84TTRR12 pKa = 11.84TIAAAAVLAVTMSGCAGGAGGGTGGGDD39 pKa = 3.16GGGGGTGYY47 pKa = 10.69EE48 pKa = 4.35YY49 pKa = 10.89GASPDD54 pKa = 3.67EE55 pKa = 4.36IKK57 pKa = 10.93AAFADD62 pKa = 4.06VDD64 pKa = 4.36PLTITYY70 pKa = 9.79QPSAQSSEE78 pKa = 4.71GIDD81 pKa = 3.46AYY83 pKa = 10.68RR84 pKa = 11.84AAAFIEE90 pKa = 3.96NLEE93 pKa = 4.23TLSDD97 pKa = 3.67GKK99 pKa = 9.53ITVDD103 pKa = 3.07TTYY106 pKa = 10.73GQGIAGYY113 pKa = 10.39DD114 pKa = 3.94EE115 pKa = 6.03LPDD118 pKa = 3.86ALVDD122 pKa = 3.72GRR124 pKa = 11.84VDD126 pKa = 3.04IAYY129 pKa = 7.33MLPIYY134 pKa = 10.54QPDD137 pKa = 3.6QFPVFHH143 pKa = 6.76AWVGGTTLTGTSPLVDD159 pKa = 3.78EE160 pKa = 5.29LAASAAIGQLTWEE173 pKa = 4.79DD174 pKa = 3.84EE175 pKa = 4.25NLLSEE180 pKa = 5.07FRR182 pKa = 11.84DD183 pKa = 3.6QGIEE187 pKa = 3.7PLTPFNSAGAIMGFCGEE204 pKa = 4.15QHH206 pKa = 6.34VDD208 pKa = 3.09ASDD211 pKa = 3.22WAGGQVRR218 pKa = 11.84ASSQAQITQLNALDD232 pKa = 4.33ASPVSLEE239 pKa = 3.81YY240 pKa = 10.92TEE242 pKa = 4.53TFEE245 pKa = 4.33ALQRR249 pKa = 11.84GTIDD253 pKa = 3.44CTMSAALAAEE263 pKa = 4.21TAGVLEE269 pKa = 4.54VAPHH273 pKa = 5.52ATYY276 pKa = 8.11TTDD279 pKa = 2.74VTFARR284 pKa = 11.84GPGGVYY290 pKa = 10.23AGSAWEE296 pKa = 4.08SWPLAVQQLVFDD308 pKa = 4.02SMEE311 pKa = 4.53DD312 pKa = 3.11EE313 pKa = 4.34FTQARR318 pKa = 11.84RR319 pKa = 11.84GDD321 pKa = 3.41LDD323 pKa = 3.84ANYY326 pKa = 9.97VAAEE330 pKa = 4.17QIRR333 pKa = 11.84EE334 pKa = 4.03QGGSFSEE341 pKa = 4.3MDD343 pKa = 4.04PDD345 pKa = 3.86AQSALLDD352 pKa = 3.8ASQGIVQDD360 pKa = 4.06AAAEE364 pKa = 4.1GHH366 pKa = 6.56LPEE369 pKa = 4.8GTVDD373 pKa = 5.09AIPAAVEE380 pKa = 3.97EE381 pKa = 4.31WRR383 pKa = 11.84GTVEE387 pKa = 3.64EE388 pKa = 5.26LGYY391 pKa = 10.82EE392 pKa = 4.18DD393 pKa = 4.15QGTFADD399 pKa = 3.88YY400 pKa = 11.28DD401 pKa = 3.33EE402 pKa = 5.33WYY404 pKa = 10.33PMDD407 pKa = 4.46DD408 pKa = 3.84EE409 pKa = 5.58EE410 pKa = 4.56YY411 pKa = 9.71LAPVGSTYY419 pKa = 10.4FEE421 pKa = 4.13EE422 pKa = 4.45VMLPHH427 pKa = 6.96RR428 pKa = 11.84PSS430 pKa = 2.95
MM1 pKa = 7.28KK2 pKa = 9.74PSRR5 pKa = 11.84ISPRR9 pKa = 11.84TTRR12 pKa = 11.84TIAAAAVLAVTMSGCAGGAGGGTGGGDD39 pKa = 3.16GGGGGTGYY47 pKa = 10.69EE48 pKa = 4.35YY49 pKa = 10.89GASPDD54 pKa = 3.67EE55 pKa = 4.36IKK57 pKa = 10.93AAFADD62 pKa = 4.06VDD64 pKa = 4.36PLTITYY70 pKa = 9.79QPSAQSSEE78 pKa = 4.71GIDD81 pKa = 3.46AYY83 pKa = 10.68RR84 pKa = 11.84AAAFIEE90 pKa = 3.96NLEE93 pKa = 4.23TLSDD97 pKa = 3.67GKK99 pKa = 9.53ITVDD103 pKa = 3.07TTYY106 pKa = 10.73GQGIAGYY113 pKa = 10.39DD114 pKa = 3.94EE115 pKa = 6.03LPDD118 pKa = 3.86ALVDD122 pKa = 3.72GRR124 pKa = 11.84VDD126 pKa = 3.04IAYY129 pKa = 7.33MLPIYY134 pKa = 10.54QPDD137 pKa = 3.6QFPVFHH143 pKa = 6.76AWVGGTTLTGTSPLVDD159 pKa = 3.78EE160 pKa = 5.29LAASAAIGQLTWEE173 pKa = 4.79DD174 pKa = 3.84EE175 pKa = 4.25NLLSEE180 pKa = 5.07FRR182 pKa = 11.84DD183 pKa = 3.6QGIEE187 pKa = 3.7PLTPFNSAGAIMGFCGEE204 pKa = 4.15QHH206 pKa = 6.34VDD208 pKa = 3.09ASDD211 pKa = 3.22WAGGQVRR218 pKa = 11.84ASSQAQITQLNALDD232 pKa = 4.33ASPVSLEE239 pKa = 3.81YY240 pKa = 10.92TEE242 pKa = 4.53TFEE245 pKa = 4.33ALQRR249 pKa = 11.84GTIDD253 pKa = 3.44CTMSAALAAEE263 pKa = 4.21TAGVLEE269 pKa = 4.54VAPHH273 pKa = 5.52ATYY276 pKa = 8.11TTDD279 pKa = 2.74VTFARR284 pKa = 11.84GPGGVYY290 pKa = 10.23AGSAWEE296 pKa = 4.08SWPLAVQQLVFDD308 pKa = 4.02SMEE311 pKa = 4.53DD312 pKa = 3.11EE313 pKa = 4.34FTQARR318 pKa = 11.84RR319 pKa = 11.84GDD321 pKa = 3.41LDD323 pKa = 3.84ANYY326 pKa = 9.97VAAEE330 pKa = 4.17QIRR333 pKa = 11.84EE334 pKa = 4.03QGGSFSEE341 pKa = 4.3MDD343 pKa = 4.04PDD345 pKa = 3.86AQSALLDD352 pKa = 3.8ASQGIVQDD360 pKa = 4.06AAAEE364 pKa = 4.1GHH366 pKa = 6.56LPEE369 pKa = 4.8GTVDD373 pKa = 5.09AIPAAVEE380 pKa = 3.97EE381 pKa = 4.31WRR383 pKa = 11.84GTVEE387 pKa = 3.64EE388 pKa = 5.26LGYY391 pKa = 10.82EE392 pKa = 4.18DD393 pKa = 4.15QGTFADD399 pKa = 3.88YY400 pKa = 11.28DD401 pKa = 3.33EE402 pKa = 5.33WYY404 pKa = 10.33PMDD407 pKa = 4.46DD408 pKa = 3.84EE409 pKa = 5.58EE410 pKa = 4.56YY411 pKa = 9.71LAPVGSTYY419 pKa = 10.4FEE421 pKa = 4.13EE422 pKa = 4.45VMLPHH427 pKa = 6.96RR428 pKa = 11.84PSS430 pKa = 2.95
Molecular weight: 45.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1X6WY65|A0A1X6WY65_9MICO Proposed peptidoglycan lipid II flippase MurJ OS=Brevibacterium yomogidense OX=946573 GN=FM105_02150 PE=4 SV=1
MM1 pKa = 7.56SKK3 pKa = 8.12HH4 pKa = 5.4TFQPNNRR11 pKa = 11.84KK12 pKa = 8.98RR13 pKa = 11.84AKK15 pKa = 8.92THH17 pKa = 5.64GFRR20 pKa = 11.84QRR22 pKa = 11.84MSTRR26 pKa = 11.84AGRR29 pKa = 11.84AILASRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84RR41 pKa = 11.84ALSAA45 pKa = 3.79
MM1 pKa = 7.56SKK3 pKa = 8.12HH4 pKa = 5.4TFQPNNRR11 pKa = 11.84KK12 pKa = 8.98RR13 pKa = 11.84AKK15 pKa = 8.92THH17 pKa = 5.64GFRR20 pKa = 11.84QRR22 pKa = 11.84MSTRR26 pKa = 11.84AGRR29 pKa = 11.84AILASRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GRR40 pKa = 11.84RR41 pKa = 11.84ALSAA45 pKa = 3.79
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
977191 |
37 |
1841 |
340.7 |
36.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.021 ± 0.075 | 0.549 ± 0.01 |
6.813 ± 0.054 | 6.108 ± 0.048 |
3.057 ± 0.027 | 9.165 ± 0.046 |
2.184 ± 0.025 | 4.26 ± 0.034 |
1.891 ± 0.029 | 9.614 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.041 ± 0.018 | 1.763 ± 0.024 |
5.381 ± 0.031 | 2.891 ± 0.025 |
7.115 ± 0.058 | 6.102 ± 0.035 |
6.233 ± 0.033 | 8.656 ± 0.044 |
1.312 ± 0.018 | 1.844 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
