
Changjiang tombus-like virus 16
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.48
Get precalculated fractions of proteins
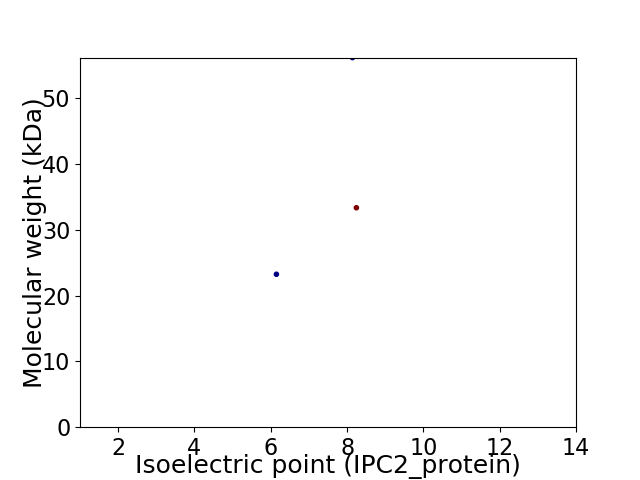
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG49|A0A1L3KG49_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 16 OX=1922809 PE=4 SV=1
MM1 pKa = 7.39ATFDD5 pKa = 3.66LGAAEE10 pKa = 4.25RR11 pKa = 11.84QLDD14 pKa = 3.8GAIGVISKK22 pKa = 9.93GFKK25 pKa = 10.12LGVMVCGVCTAGYY38 pKa = 9.18LAYY41 pKa = 10.2RR42 pKa = 11.84YY43 pKa = 9.9VPRR46 pKa = 11.84LWRR49 pKa = 11.84LMEE52 pKa = 4.24VMDD55 pKa = 4.97HH56 pKa = 6.47SLTGKK61 pKa = 9.01EE62 pKa = 3.7RR63 pKa = 11.84AMEE66 pKa = 4.18LQSKK70 pKa = 8.97AVEE73 pKa = 4.11YY74 pKa = 11.21VPDD77 pKa = 3.84EE78 pKa = 4.49DD79 pKa = 4.2TDD81 pKa = 3.8EE82 pKa = 4.67EE83 pKa = 4.33EE84 pKa = 4.96ADD86 pKa = 4.66VGAEE90 pKa = 4.78DD91 pKa = 4.44NDD93 pKa = 3.85APPGLGRR100 pKa = 11.84RR101 pKa = 11.84GPAARR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84ARR110 pKa = 11.84KK111 pKa = 8.96EE112 pKa = 3.27KK113 pKa = 8.63HH114 pKa = 5.45TGMDD118 pKa = 4.0GVPLTNYY125 pKa = 8.41YY126 pKa = 11.09ASIVSEE132 pKa = 3.93ARR134 pKa = 11.84VHH136 pKa = 5.96YY137 pKa = 9.72GYY139 pKa = 10.75KK140 pKa = 10.47GYY142 pKa = 10.91SSYY145 pKa = 11.44NAALARR151 pKa = 11.84AYY153 pKa = 8.5MVRR156 pKa = 11.84LMTEE160 pKa = 3.69HH161 pKa = 6.7NVRR164 pKa = 11.84KK165 pKa = 9.79AHH167 pKa = 5.27MLEE170 pKa = 4.04NLEE173 pKa = 4.53DD174 pKa = 3.5MVNAVFAVSAVDD186 pKa = 4.5RR187 pKa = 11.84RR188 pKa = 11.84AQLEE192 pKa = 4.25RR193 pKa = 11.84EE194 pKa = 4.12AGLLFGWMRR203 pKa = 11.84RR204 pKa = 11.84AQNCC208 pKa = 3.24
MM1 pKa = 7.39ATFDD5 pKa = 3.66LGAAEE10 pKa = 4.25RR11 pKa = 11.84QLDD14 pKa = 3.8GAIGVISKK22 pKa = 9.93GFKK25 pKa = 10.12LGVMVCGVCTAGYY38 pKa = 9.18LAYY41 pKa = 10.2RR42 pKa = 11.84YY43 pKa = 9.9VPRR46 pKa = 11.84LWRR49 pKa = 11.84LMEE52 pKa = 4.24VMDD55 pKa = 4.97HH56 pKa = 6.47SLTGKK61 pKa = 9.01EE62 pKa = 3.7RR63 pKa = 11.84AMEE66 pKa = 4.18LQSKK70 pKa = 8.97AVEE73 pKa = 4.11YY74 pKa = 11.21VPDD77 pKa = 3.84EE78 pKa = 4.49DD79 pKa = 4.2TDD81 pKa = 3.8EE82 pKa = 4.67EE83 pKa = 4.33EE84 pKa = 4.96ADD86 pKa = 4.66VGAEE90 pKa = 4.78DD91 pKa = 4.44NDD93 pKa = 3.85APPGLGRR100 pKa = 11.84RR101 pKa = 11.84GPAARR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84ARR110 pKa = 11.84KK111 pKa = 8.96EE112 pKa = 3.27KK113 pKa = 8.63HH114 pKa = 5.45TGMDD118 pKa = 4.0GVPLTNYY125 pKa = 8.41YY126 pKa = 11.09ASIVSEE132 pKa = 3.93ARR134 pKa = 11.84VHH136 pKa = 5.96YY137 pKa = 9.72GYY139 pKa = 10.75KK140 pKa = 10.47GYY142 pKa = 10.91SSYY145 pKa = 11.44NAALARR151 pKa = 11.84AYY153 pKa = 8.5MVRR156 pKa = 11.84LMTEE160 pKa = 3.69HH161 pKa = 6.7NVRR164 pKa = 11.84KK165 pKa = 9.79AHH167 pKa = 5.27MLEE170 pKa = 4.04NLEE173 pKa = 4.53DD174 pKa = 3.5MVNAVFAVSAVDD186 pKa = 4.5RR187 pKa = 11.84RR188 pKa = 11.84AQLEE192 pKa = 4.25RR193 pKa = 11.84EE194 pKa = 4.12AGLLFGWMRR203 pKa = 11.84RR204 pKa = 11.84AQNCC208 pKa = 3.24
Molecular weight: 23.23 kDa
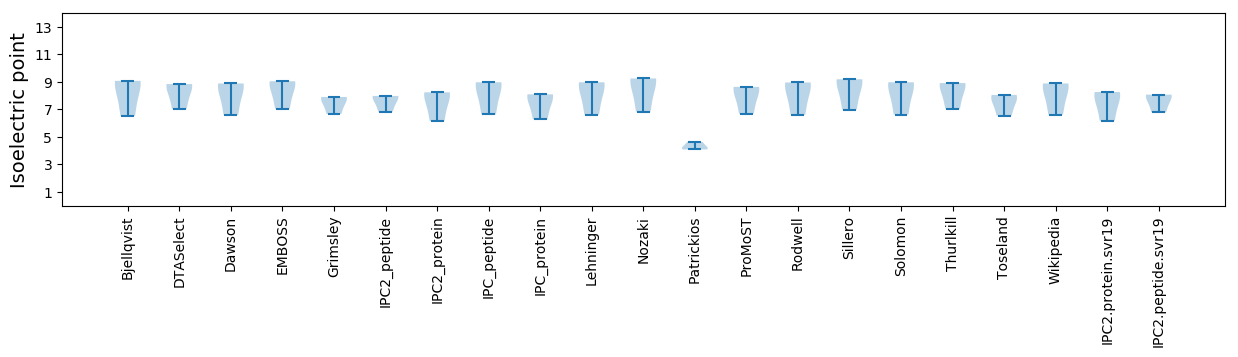
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG49|A0A1L3KG49_9VIRU Uncharacterized protein OS=Changjiang tombus-like virus 16 OX=1922809 PE=4 SV=1
MM1 pKa = 7.73AKK3 pKa = 10.11NKK5 pKa = 10.26KK6 pKa = 10.1SDD8 pKa = 3.59VKK10 pKa = 10.57KK11 pKa = 10.96SKK13 pKa = 10.89AGTSKK18 pKa = 10.05YY19 pKa = 8.71QKK21 pKa = 10.14KK22 pKa = 10.19AFGMYY27 pKa = 9.6KK28 pKa = 9.92QIEE31 pKa = 4.63AGTLDD36 pKa = 3.59RR37 pKa = 11.84AALEE41 pKa = 4.13AAVMYY46 pKa = 10.41SDD48 pKa = 4.06PCGADD53 pKa = 3.33LVPTVYY59 pKa = 10.41PGDD62 pKa = 3.48AGYY65 pKa = 10.48INRR68 pKa = 11.84FNTINTVGTVAGQTATVMIWRR89 pKa = 11.84PGTQGCVEE97 pKa = 4.32YY98 pKa = 10.67GVADD102 pKa = 3.74GGVGVVTAYY111 pKa = 7.93NTNFPGLTFLNTNASKK127 pKa = 9.32TRR129 pKa = 11.84CGAFCATVRR138 pKa = 11.84PVAAPNTCTGTIYY151 pKa = 10.64FGVVNARR158 pKa = 11.84AVRR161 pKa = 11.84SGLNLSPNAMIPLCSEE177 pKa = 4.29SVSASQALMAPLEE190 pKa = 4.43VRR192 pKa = 11.84WSPGDD197 pKa = 3.19FDD199 pKa = 4.46SRR201 pKa = 11.84YY202 pKa = 10.44SEE204 pKa = 4.21VNNPNVAADD213 pKa = 3.87DD214 pKa = 3.8SDD216 pKa = 4.56YY217 pKa = 11.84NCLLVVAIGLPAATGIQYY235 pKa = 10.33RR236 pKa = 11.84ATSIMEE242 pKa = 3.99WCPNSNLGIANDD254 pKa = 3.67ATRR257 pKa = 11.84VKK259 pKa = 10.49PSACDD264 pKa = 3.49KK265 pKa = 10.64EE266 pKa = 4.82CVLNFLKK273 pKa = 10.82KK274 pKa = 9.68KK275 pKa = 10.49DD276 pKa = 3.94KK277 pKa = 10.68NWWWSLGNKK286 pKa = 6.09TLKK289 pKa = 9.57VAKK292 pKa = 8.81TVASGYY298 pKa = 9.92YY299 pKa = 9.54SGGAIGALGSLTKK312 pKa = 10.52FMM314 pKa = 5.62
MM1 pKa = 7.73AKK3 pKa = 10.11NKK5 pKa = 10.26KK6 pKa = 10.1SDD8 pKa = 3.59VKK10 pKa = 10.57KK11 pKa = 10.96SKK13 pKa = 10.89AGTSKK18 pKa = 10.05YY19 pKa = 8.71QKK21 pKa = 10.14KK22 pKa = 10.19AFGMYY27 pKa = 9.6KK28 pKa = 9.92QIEE31 pKa = 4.63AGTLDD36 pKa = 3.59RR37 pKa = 11.84AALEE41 pKa = 4.13AAVMYY46 pKa = 10.41SDD48 pKa = 4.06PCGADD53 pKa = 3.33LVPTVYY59 pKa = 10.41PGDD62 pKa = 3.48AGYY65 pKa = 10.48INRR68 pKa = 11.84FNTINTVGTVAGQTATVMIWRR89 pKa = 11.84PGTQGCVEE97 pKa = 4.32YY98 pKa = 10.67GVADD102 pKa = 3.74GGVGVVTAYY111 pKa = 7.93NTNFPGLTFLNTNASKK127 pKa = 9.32TRR129 pKa = 11.84CGAFCATVRR138 pKa = 11.84PVAAPNTCTGTIYY151 pKa = 10.64FGVVNARR158 pKa = 11.84AVRR161 pKa = 11.84SGLNLSPNAMIPLCSEE177 pKa = 4.29SVSASQALMAPLEE190 pKa = 4.43VRR192 pKa = 11.84WSPGDD197 pKa = 3.19FDD199 pKa = 4.46SRR201 pKa = 11.84YY202 pKa = 10.44SEE204 pKa = 4.21VNNPNVAADD213 pKa = 3.87DD214 pKa = 3.8SDD216 pKa = 4.56YY217 pKa = 11.84NCLLVVAIGLPAATGIQYY235 pKa = 10.33RR236 pKa = 11.84ATSIMEE242 pKa = 3.99WCPNSNLGIANDD254 pKa = 3.67ATRR257 pKa = 11.84VKK259 pKa = 10.49PSACDD264 pKa = 3.49KK265 pKa = 10.64EE266 pKa = 4.82CVLNFLKK273 pKa = 10.82KK274 pKa = 9.68KK275 pKa = 10.49DD276 pKa = 3.94KK277 pKa = 10.68NWWWSLGNKK286 pKa = 6.09TLKK289 pKa = 9.57VAKK292 pKa = 8.81TVASGYY298 pKa = 9.92YY299 pKa = 9.54SGGAIGALGSLTKK312 pKa = 10.52FMM314 pKa = 5.62
Molecular weight: 33.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1021 |
208 |
499 |
340.3 |
37.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.696 ± 1.605 | 2.351 ± 0.331 |
4.995 ± 0.243 | 5.387 ± 1.091 |
3.134 ± 0.415 | 8.227 ± 0.459 |
1.567 ± 0.561 | 3.232 ± 0.503 |
5.093 ± 0.585 | 9.403 ± 1.371 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.036 ± 0.623 | 4.016 ± 1.104 |
4.505 ± 0.454 | 3.232 ± 0.761 |
7.15 ± 1.317 | 6.17 ± 0.641 |
5.191 ± 0.894 | 8.423 ± 0.115 |
1.469 ± 0.179 | 3.722 ± 0.554 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
