
Momordica charantia associated gemycircularvirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus mocha1
Average proteome isoelectric point is 7.33
Get precalculated fractions of proteins
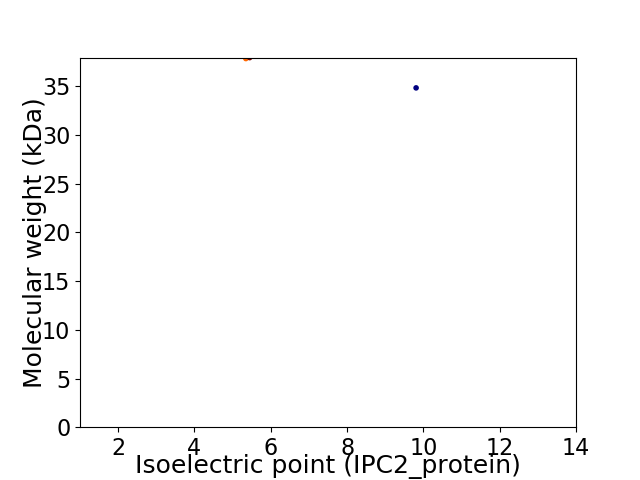
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A345S7U9|A0A345S7U9_9VIRU Replication-associated protein OS=Momordica charantia associated gemycircularvirus OX=2291613 PE=3 SV=1
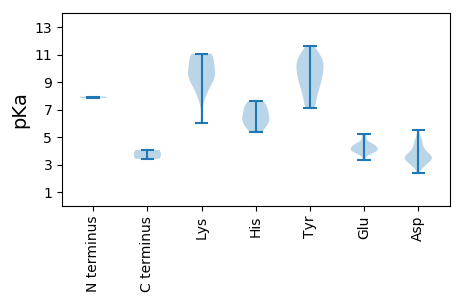
MM1 pKa = 7.84PSFVCNFRR9 pKa = 11.84YY10 pKa = 10.28GLITYY15 pKa = 7.3PQCDD19 pKa = 3.52GLDD22 pKa = 3.43PFRR25 pKa = 11.84VMEE28 pKa = 4.46RR29 pKa = 11.84FSSLGAEE36 pKa = 4.35CIIGRR41 pKa = 11.84EE42 pKa = 3.83RR43 pKa = 11.84HH44 pKa = 5.72EE45 pKa = 5.01DD46 pKa = 3.46GGLHH50 pKa = 5.74LHH52 pKa = 6.24VFADD56 pKa = 4.7FGRR59 pKa = 11.84KK60 pKa = 8.14FRR62 pKa = 11.84SRR64 pKa = 11.84KK65 pKa = 7.79TDD67 pKa = 2.98VFDD70 pKa = 3.81VDD72 pKa = 3.65GRR74 pKa = 11.84HH75 pKa = 6.28PNVEE79 pKa = 4.04PSKK82 pKa = 9.22GTPEE86 pKa = 3.79KK87 pKa = 10.85GYY89 pKa = 10.88DD90 pKa = 3.47YY91 pKa = 10.69AIKK94 pKa = 10.83DD95 pKa = 3.47GDD97 pKa = 4.33VVCGGLGRR105 pKa = 11.84PEE107 pKa = 3.97PSGGGDD113 pKa = 3.43GASSAKK119 pKa = 8.61WARR122 pKa = 11.84ITQAEE127 pKa = 4.11NRR129 pKa = 11.84DD130 pKa = 3.9EE131 pKa = 4.33FWEE134 pKa = 4.38LVHH137 pKa = 7.01EE138 pKa = 5.14LDD140 pKa = 4.73PKK142 pKa = 10.9AAACSFNALSKK153 pKa = 9.33YY154 pKa = 10.43ADD156 pKa = 3.0WRR158 pKa = 11.84FAEE161 pKa = 4.33VPPEE165 pKa = 3.98YY166 pKa = 10.36EE167 pKa = 3.34HH168 pKa = 7.5DD169 pKa = 3.61GRR171 pKa = 11.84IEE173 pKa = 4.01FVPGDD178 pKa = 3.59ADD180 pKa = 4.72GRR182 pKa = 11.84ADD184 pKa = 3.28WVSQSGIRR192 pKa = 11.84LAEE195 pKa = 4.12PFLAGTRR202 pKa = 11.84VKK204 pKa = 10.87SLVLWGKK211 pKa = 10.19SRR213 pKa = 11.84TGKK216 pKa = 6.05TTWARR221 pKa = 11.84SLGPHH226 pKa = 6.31VYY228 pKa = 10.18CVGLVSGTEE237 pKa = 4.02CAKK240 pKa = 10.98GLDD243 pKa = 3.16ARR245 pKa = 11.84YY246 pKa = 9.64AVFDD250 pKa = 5.19DD251 pKa = 3.36IRR253 pKa = 11.84GGLGFFHH260 pKa = 7.61GYY262 pKa = 9.93KK263 pKa = 9.57EE264 pKa = 3.99WLGAQPHH271 pKa = 5.37VPIKK275 pKa = 10.13EE276 pKa = 4.14LYY278 pKa = 9.69RR279 pKa = 11.84EE280 pKa = 4.04PRR282 pKa = 11.84YY283 pKa = 9.43MRR285 pKa = 11.84WNKK288 pKa = 9.74PSIWVCNRR296 pKa = 11.84DD297 pKa = 3.62PRR299 pKa = 11.84LEE301 pKa = 4.31AYY303 pKa = 8.55PPNATPDD310 pKa = 3.91WEE312 pKa = 4.27WMDD315 pKa = 4.74DD316 pKa = 3.47NVDD319 pKa = 4.4FIEE322 pKa = 4.46VTDD325 pKa = 4.0SLIASISHH333 pKa = 7.17ASTEE337 pKa = 4.06
MM1 pKa = 7.84PSFVCNFRR9 pKa = 11.84YY10 pKa = 10.28GLITYY15 pKa = 7.3PQCDD19 pKa = 3.52GLDD22 pKa = 3.43PFRR25 pKa = 11.84VMEE28 pKa = 4.46RR29 pKa = 11.84FSSLGAEE36 pKa = 4.35CIIGRR41 pKa = 11.84EE42 pKa = 3.83RR43 pKa = 11.84HH44 pKa = 5.72EE45 pKa = 5.01DD46 pKa = 3.46GGLHH50 pKa = 5.74LHH52 pKa = 6.24VFADD56 pKa = 4.7FGRR59 pKa = 11.84KK60 pKa = 8.14FRR62 pKa = 11.84SRR64 pKa = 11.84KK65 pKa = 7.79TDD67 pKa = 2.98VFDD70 pKa = 3.81VDD72 pKa = 3.65GRR74 pKa = 11.84HH75 pKa = 6.28PNVEE79 pKa = 4.04PSKK82 pKa = 9.22GTPEE86 pKa = 3.79KK87 pKa = 10.85GYY89 pKa = 10.88DD90 pKa = 3.47YY91 pKa = 10.69AIKK94 pKa = 10.83DD95 pKa = 3.47GDD97 pKa = 4.33VVCGGLGRR105 pKa = 11.84PEE107 pKa = 3.97PSGGGDD113 pKa = 3.43GASSAKK119 pKa = 8.61WARR122 pKa = 11.84ITQAEE127 pKa = 4.11NRR129 pKa = 11.84DD130 pKa = 3.9EE131 pKa = 4.33FWEE134 pKa = 4.38LVHH137 pKa = 7.01EE138 pKa = 5.14LDD140 pKa = 4.73PKK142 pKa = 10.9AAACSFNALSKK153 pKa = 9.33YY154 pKa = 10.43ADD156 pKa = 3.0WRR158 pKa = 11.84FAEE161 pKa = 4.33VPPEE165 pKa = 3.98YY166 pKa = 10.36EE167 pKa = 3.34HH168 pKa = 7.5DD169 pKa = 3.61GRR171 pKa = 11.84IEE173 pKa = 4.01FVPGDD178 pKa = 3.59ADD180 pKa = 4.72GRR182 pKa = 11.84ADD184 pKa = 3.28WVSQSGIRR192 pKa = 11.84LAEE195 pKa = 4.12PFLAGTRR202 pKa = 11.84VKK204 pKa = 10.87SLVLWGKK211 pKa = 10.19SRR213 pKa = 11.84TGKK216 pKa = 6.05TTWARR221 pKa = 11.84SLGPHH226 pKa = 6.31VYY228 pKa = 10.18CVGLVSGTEE237 pKa = 4.02CAKK240 pKa = 10.98GLDD243 pKa = 3.16ARR245 pKa = 11.84YY246 pKa = 9.64AVFDD250 pKa = 5.19DD251 pKa = 3.36IRR253 pKa = 11.84GGLGFFHH260 pKa = 7.61GYY262 pKa = 9.93KK263 pKa = 9.57EE264 pKa = 3.99WLGAQPHH271 pKa = 5.37VPIKK275 pKa = 10.13EE276 pKa = 4.14LYY278 pKa = 9.69RR279 pKa = 11.84EE280 pKa = 4.04PRR282 pKa = 11.84YY283 pKa = 9.43MRR285 pKa = 11.84WNKK288 pKa = 9.74PSIWVCNRR296 pKa = 11.84DD297 pKa = 3.62PRR299 pKa = 11.84LEE301 pKa = 4.31AYY303 pKa = 8.55PPNATPDD310 pKa = 3.91WEE312 pKa = 4.27WMDD315 pKa = 4.74DD316 pKa = 3.47NVDD319 pKa = 4.4FIEE322 pKa = 4.46VTDD325 pKa = 4.0SLIASISHH333 pKa = 7.17ASTEE337 pKa = 4.06
Molecular weight: 37.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345S7U9|A0A345S7U9_9VIRU Replication-associated protein OS=Momordica charantia associated gemycircularvirus OX=2291613 PE=3 SV=1
MM1 pKa = 7.95AYY3 pKa = 9.64RR4 pKa = 11.84RR5 pKa = 11.84SVRR8 pKa = 11.84RR9 pKa = 11.84APARR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PTRR18 pKa = 11.84KK19 pKa = 8.77SSYY22 pKa = 7.52GSRR25 pKa = 11.84YY26 pKa = 8.58RR27 pKa = 11.84RR28 pKa = 11.84SYY30 pKa = 7.13TAKK33 pKa = 10.15RR34 pKa = 11.84RR35 pKa = 11.84FTRR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 8.93RR41 pKa = 11.84PMSRR45 pKa = 11.84KK46 pKa = 9.14SLLNVTSRR54 pKa = 11.84KK55 pKa = 9.88KK56 pKa = 9.81QDD58 pKa = 2.94TMLGYY63 pKa = 11.12SNLANDD69 pKa = 4.13STDD72 pKa = 2.91GSNKK76 pKa = 8.33FHH78 pKa = 7.02SGAGILKK85 pKa = 9.23GGRR88 pKa = 11.84MYY90 pKa = 10.49IIPWIATARR99 pKa = 11.84DD100 pKa = 3.65LSPTDD105 pKa = 3.44GTLGTVAQKK114 pKa = 10.95ASRR117 pKa = 11.84TSTEE121 pKa = 3.71CYY123 pKa = 9.18MRR125 pKa = 11.84GLRR128 pKa = 11.84EE129 pKa = 4.33RR130 pKa = 11.84IQIQTNNGTSWQWRR144 pKa = 11.84RR145 pKa = 11.84ICFTFKK151 pKa = 11.06GPLLVNATTGSEE163 pKa = 3.99TQRR166 pKa = 11.84YY167 pKa = 8.48QIEE170 pKa = 4.48TNQGFARR177 pKa = 11.84VMSDD181 pKa = 3.61FALGTAADD189 pKa = 4.26PLNGVLFKK197 pKa = 11.03GLQSVDD203 pKa = 2.37WSSYY207 pKa = 7.47FTAKK211 pKa = 9.67TDD213 pKa = 3.38NSLVNIMYY221 pKa = 9.37DD222 pKa = 3.01QTRR225 pKa = 11.84IISSGNASGVMRR237 pKa = 11.84NFKK240 pKa = 9.79LWHH243 pKa = 6.22PMNKK247 pKa = 9.33NLYY250 pKa = 8.36YY251 pKa = 10.85NEE253 pKa = 4.64DD254 pKa = 3.89EE255 pKa = 4.58IGATTGTSVYY265 pKa = 9.16STSSKK270 pKa = 10.77RR271 pKa = 11.84GMGDD275 pKa = 3.32YY276 pKa = 10.78YY277 pKa = 11.59VVDD280 pKa = 5.52FILAGTGSNINDD292 pKa = 3.41LMSFEE297 pKa = 5.22PDD299 pKa = 2.85STLYY303 pKa = 8.55WHH305 pKa = 6.72EE306 pKa = 4.03RR307 pKa = 3.41
MM1 pKa = 7.95AYY3 pKa = 9.64RR4 pKa = 11.84RR5 pKa = 11.84SVRR8 pKa = 11.84RR9 pKa = 11.84APARR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84PTRR18 pKa = 11.84KK19 pKa = 8.77SSYY22 pKa = 7.52GSRR25 pKa = 11.84YY26 pKa = 8.58RR27 pKa = 11.84RR28 pKa = 11.84SYY30 pKa = 7.13TAKK33 pKa = 10.15RR34 pKa = 11.84RR35 pKa = 11.84FTRR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 8.93RR41 pKa = 11.84PMSRR45 pKa = 11.84KK46 pKa = 9.14SLLNVTSRR54 pKa = 11.84KK55 pKa = 9.88KK56 pKa = 9.81QDD58 pKa = 2.94TMLGYY63 pKa = 11.12SNLANDD69 pKa = 4.13STDD72 pKa = 2.91GSNKK76 pKa = 8.33FHH78 pKa = 7.02SGAGILKK85 pKa = 9.23GGRR88 pKa = 11.84MYY90 pKa = 10.49IIPWIATARR99 pKa = 11.84DD100 pKa = 3.65LSPTDD105 pKa = 3.44GTLGTVAQKK114 pKa = 10.95ASRR117 pKa = 11.84TSTEE121 pKa = 3.71CYY123 pKa = 9.18MRR125 pKa = 11.84GLRR128 pKa = 11.84EE129 pKa = 4.33RR130 pKa = 11.84IQIQTNNGTSWQWRR144 pKa = 11.84RR145 pKa = 11.84ICFTFKK151 pKa = 11.06GPLLVNATTGSEE163 pKa = 3.99TQRR166 pKa = 11.84YY167 pKa = 8.48QIEE170 pKa = 4.48TNQGFARR177 pKa = 11.84VMSDD181 pKa = 3.61FALGTAADD189 pKa = 4.26PLNGVLFKK197 pKa = 11.03GLQSVDD203 pKa = 2.37WSSYY207 pKa = 7.47FTAKK211 pKa = 9.67TDD213 pKa = 3.38NSLVNIMYY221 pKa = 9.37DD222 pKa = 3.01QTRR225 pKa = 11.84IISSGNASGVMRR237 pKa = 11.84NFKK240 pKa = 9.79LWHH243 pKa = 6.22PMNKK247 pKa = 9.33NLYY250 pKa = 8.36YY251 pKa = 10.85NEE253 pKa = 4.64DD254 pKa = 3.89EE255 pKa = 4.58IGATTGTSVYY265 pKa = 9.16STSSKK270 pKa = 10.77RR271 pKa = 11.84GMGDD275 pKa = 3.32YY276 pKa = 10.78YY277 pKa = 11.59VVDD280 pKa = 5.52FILAGTGSNINDD292 pKa = 3.41LMSFEE297 pKa = 5.22PDD299 pKa = 2.85STLYY303 pKa = 8.55WHH305 pKa = 6.72EE306 pKa = 4.03RR307 pKa = 3.41
Molecular weight: 34.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
644 |
307 |
337 |
322.0 |
36.35 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.988 ± 0.566 | 1.553 ± 0.639 |
6.522 ± 1.159 | 4.969 ± 1.675 |
4.348 ± 0.542 | 9.161 ± 0.722 |
2.019 ± 0.738 | 4.193 ± 0.261 |
4.658 ± 0.161 | 6.366 ± 0.105 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.329 ± 0.889 | 4.037 ± 1.294 |
4.814 ± 1.334 | 2.174 ± 0.768 |
9.006 ± 1.004 | 8.23 ± 1.555 |
6.366 ± 2.183 | 5.28 ± 0.971 |
2.64 ± 0.486 | 4.348 ± 0.612 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
