
Cryobacterium arcticum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Cryobacterium
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
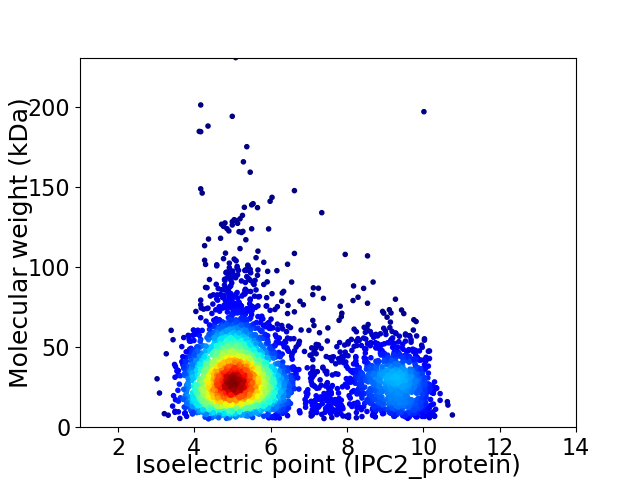
Virtual 2D-PAGE plot for 3936 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
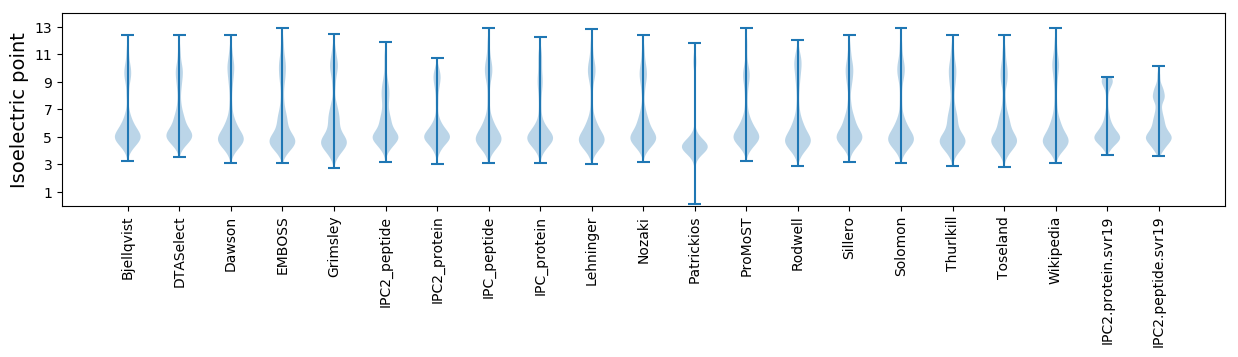
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B1BNE9|A0A1B1BNE9_9MICO Transcriptional regulator OS=Cryobacterium arcticum OX=670052 GN=PA27867_3263 PE=3 SV=1
MM1 pKa = 6.82ATKK4 pKa = 10.42RR5 pKa = 11.84HH6 pKa = 6.14PATFLGAGVAATVLFLSGCAANEE29 pKa = 4.27AGDD32 pKa = 4.31EE33 pKa = 4.15PSDD36 pKa = 4.54LAGTLDD42 pKa = 3.8GAGSSAQASAEE53 pKa = 4.2DD54 pKa = 3.5VWISQFQRR62 pKa = 11.84ANGSVTINYY71 pKa = 8.96DD72 pKa = 3.27PTGSGAGRR80 pKa = 11.84EE81 pKa = 3.99QFIGGGVNFAGSDD94 pKa = 3.48SALSDD99 pKa = 4.09DD100 pKa = 4.32EE101 pKa = 4.61LAGSFGACAPGSAAIDD117 pKa = 3.62LPVYY121 pKa = 10.27VSPLVLVYY129 pKa = 10.82NVDD132 pKa = 3.66GVDD135 pKa = 3.89SLRR138 pKa = 11.84LDD140 pKa = 3.79PQTIASIYY148 pKa = 10.16SGAITRR154 pKa = 11.84WNDD157 pKa = 3.13PALVALNPDD166 pKa = 3.31AGLPDD171 pKa = 3.77EE172 pKa = 5.66PISAVHH178 pKa = 6.78RR179 pKa = 11.84SDD181 pKa = 5.84DD182 pKa = 3.99SGTTKK187 pKa = 10.73NFADD191 pKa = 3.8YY192 pKa = 10.6LYY194 pKa = 11.1QNVPDD199 pKa = 3.39VWTNEE204 pKa = 3.91PADD207 pKa = 3.77AFPYY211 pKa = 9.94PGEE214 pKa = 4.25GAQGNSGVVSAVANGRR230 pKa = 11.84NTIGYY235 pKa = 9.71ADD237 pKa = 4.09ASRR240 pKa = 11.84VGDD243 pKa = 3.86LSVAALKK250 pKa = 10.68VGNDD254 pKa = 3.32FVDD257 pKa = 3.94YY258 pKa = 9.63STEE261 pKa = 3.54AAAAIIDD268 pKa = 4.1EE269 pKa = 4.67SPLVARR275 pKa = 11.84DD276 pKa = 3.69NPNDD280 pKa = 3.6LVVDD284 pKa = 3.73VDD286 pKa = 4.06RR287 pKa = 11.84TSTAAGVYY295 pKa = 9.4PLVLVSYY302 pKa = 10.12LIACEE307 pKa = 4.35EE308 pKa = 4.27YY309 pKa = 10.26PDD311 pKa = 4.51ADD313 pKa = 3.53QAEE316 pKa = 4.77LVSAYY321 pKa = 10.0LGYY324 pKa = 10.25VASPEE329 pKa = 4.25GQNAAAEE336 pKa = 4.57SAGSAPLSAEE346 pKa = 4.07FSARR350 pKa = 11.84VLDD353 pKa = 5.24AIDD356 pKa = 4.07SIRR359 pKa = 3.71
MM1 pKa = 6.82ATKK4 pKa = 10.42RR5 pKa = 11.84HH6 pKa = 6.14PATFLGAGVAATVLFLSGCAANEE29 pKa = 4.27AGDD32 pKa = 4.31EE33 pKa = 4.15PSDD36 pKa = 4.54LAGTLDD42 pKa = 3.8GAGSSAQASAEE53 pKa = 4.2DD54 pKa = 3.5VWISQFQRR62 pKa = 11.84ANGSVTINYY71 pKa = 8.96DD72 pKa = 3.27PTGSGAGRR80 pKa = 11.84EE81 pKa = 3.99QFIGGGVNFAGSDD94 pKa = 3.48SALSDD99 pKa = 4.09DD100 pKa = 4.32EE101 pKa = 4.61LAGSFGACAPGSAAIDD117 pKa = 3.62LPVYY121 pKa = 10.27VSPLVLVYY129 pKa = 10.82NVDD132 pKa = 3.66GVDD135 pKa = 3.89SLRR138 pKa = 11.84LDD140 pKa = 3.79PQTIASIYY148 pKa = 10.16SGAITRR154 pKa = 11.84WNDD157 pKa = 3.13PALVALNPDD166 pKa = 3.31AGLPDD171 pKa = 3.77EE172 pKa = 5.66PISAVHH178 pKa = 6.78RR179 pKa = 11.84SDD181 pKa = 5.84DD182 pKa = 3.99SGTTKK187 pKa = 10.73NFADD191 pKa = 3.8YY192 pKa = 10.6LYY194 pKa = 11.1QNVPDD199 pKa = 3.39VWTNEE204 pKa = 3.91PADD207 pKa = 3.77AFPYY211 pKa = 9.94PGEE214 pKa = 4.25GAQGNSGVVSAVANGRR230 pKa = 11.84NTIGYY235 pKa = 9.71ADD237 pKa = 4.09ASRR240 pKa = 11.84VGDD243 pKa = 3.86LSVAALKK250 pKa = 10.68VGNDD254 pKa = 3.32FVDD257 pKa = 3.94YY258 pKa = 9.63STEE261 pKa = 3.54AAAAIIDD268 pKa = 4.1EE269 pKa = 4.67SPLVARR275 pKa = 11.84DD276 pKa = 3.69NPNDD280 pKa = 3.6LVVDD284 pKa = 3.73VDD286 pKa = 4.06RR287 pKa = 11.84TSTAAGVYY295 pKa = 9.4PLVLVSYY302 pKa = 10.12LIACEE307 pKa = 4.35EE308 pKa = 4.27YY309 pKa = 10.26PDD311 pKa = 4.51ADD313 pKa = 3.53QAEE316 pKa = 4.77LVSAYY321 pKa = 10.0LGYY324 pKa = 10.25VASPEE329 pKa = 4.25GQNAAAEE336 pKa = 4.57SAGSAPLSAEE346 pKa = 4.07FSARR350 pKa = 11.84VLDD353 pKa = 5.24AIDD356 pKa = 4.07SIRR359 pKa = 3.71
Molecular weight: 36.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B1BQL1|A0A1B1BQL1_9MICO Uncharacterized protein OS=Cryobacterium arcticum OX=670052 GN=PA27867_3937 PE=4 SV=1
MM1 pKa = 7.44SANSLKK7 pKa = 10.63RR8 pKa = 11.84KK9 pKa = 9.57LLLIKK14 pKa = 9.94QLHH17 pKa = 5.92KK18 pKa = 10.71RR19 pKa = 11.84GPTDD23 pKa = 3.22PKK25 pKa = 10.24EE26 pKa = 3.82IRR28 pKa = 11.84RR29 pKa = 11.84LLSCEE34 pKa = 3.85RR35 pKa = 11.84LRR37 pKa = 11.84LRR39 pKa = 11.84HH40 pKa = 4.86NTHH43 pKa = 7.07RR44 pKa = 11.84KK45 pKa = 8.45PPLHH49 pKa = 5.96GVNYY53 pKa = 9.86LQEE56 pKa = 5.1HH57 pKa = 6.49PMNLFWDD64 pKa = 4.19LDD66 pKa = 3.86LVTRR70 pKa = 11.84CGAGEE75 pKa = 4.21KK76 pKa = 8.18VTRR79 pKa = 11.84DD80 pKa = 3.21RR81 pKa = 11.84GLSTGSLVRR90 pKa = 11.84RR91 pKa = 11.84EE92 pKa = 4.52KK93 pKa = 10.28IVHH96 pKa = 6.68PGDD99 pKa = 3.33TSQVVAYY106 pKa = 10.11VCLLAQCHH114 pKa = 5.12RR115 pKa = 11.84VAHH118 pKa = 6.17NYY120 pKa = 10.74SLFARR125 pKa = 11.84NVTHH129 pKa = 7.41LRR131 pKa = 11.84IFF133 pKa = 3.73
MM1 pKa = 7.44SANSLKK7 pKa = 10.63RR8 pKa = 11.84KK9 pKa = 9.57LLLIKK14 pKa = 9.94QLHH17 pKa = 5.92KK18 pKa = 10.71RR19 pKa = 11.84GPTDD23 pKa = 3.22PKK25 pKa = 10.24EE26 pKa = 3.82IRR28 pKa = 11.84RR29 pKa = 11.84LLSCEE34 pKa = 3.85RR35 pKa = 11.84LRR37 pKa = 11.84LRR39 pKa = 11.84HH40 pKa = 4.86NTHH43 pKa = 7.07RR44 pKa = 11.84KK45 pKa = 8.45PPLHH49 pKa = 5.96GVNYY53 pKa = 9.86LQEE56 pKa = 5.1HH57 pKa = 6.49PMNLFWDD64 pKa = 4.19LDD66 pKa = 3.86LVTRR70 pKa = 11.84CGAGEE75 pKa = 4.21KK76 pKa = 8.18VTRR79 pKa = 11.84DD80 pKa = 3.21RR81 pKa = 11.84GLSTGSLVRR90 pKa = 11.84RR91 pKa = 11.84EE92 pKa = 4.52KK93 pKa = 10.28IVHH96 pKa = 6.68PGDD99 pKa = 3.33TSQVVAYY106 pKa = 10.11VCLLAQCHH114 pKa = 5.12RR115 pKa = 11.84VAHH118 pKa = 6.17NYY120 pKa = 10.74SLFARR125 pKa = 11.84NVTHH129 pKa = 7.41LRR131 pKa = 11.84IFF133 pKa = 3.73
Molecular weight: 15.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1284498 |
50 |
2228 |
326.3 |
34.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.351 ± 0.055 | 0.508 ± 0.009 |
5.989 ± 0.037 | 5.045 ± 0.041 |
3.184 ± 0.023 | 8.885 ± 0.035 |
1.924 ± 0.021 | 4.58 ± 0.028 |
2.035 ± 0.028 | 10.531 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.766 ± 0.016 | 2.238 ± 0.021 |
5.516 ± 0.028 | 2.883 ± 0.021 |
6.627 ± 0.046 | 5.952 ± 0.03 |
6.685 ± 0.039 | 8.796 ± 0.037 |
1.469 ± 0.02 | 2.038 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
