
Caprine arthritis encephalitis virus (CAEV)
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Orthoretrovirinae; Lentivirus
Average proteome isoelectric point is 8.49
Get precalculated fractions of proteins
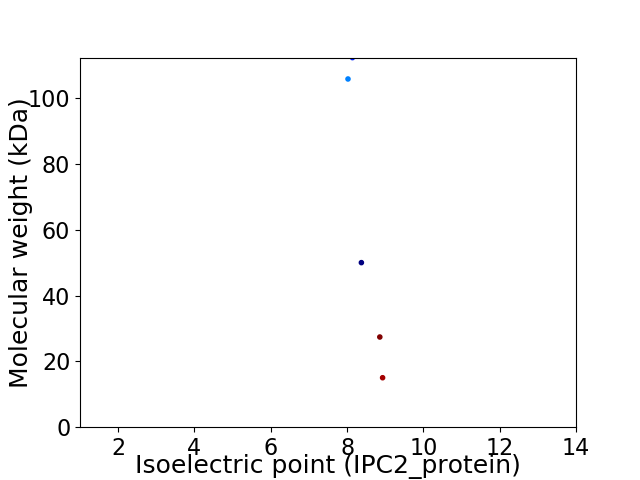
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C9DI74|C9DI74_CAEV Rev protein OS=Caprine arthritis encephalitis virus OX=11660 GN=rev PE=4 SV=1
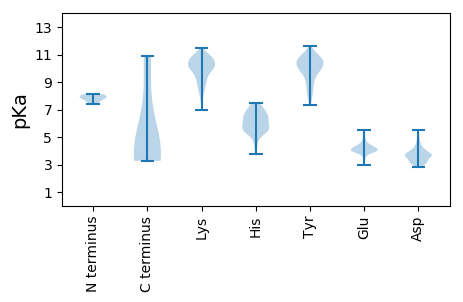
MM1 pKa = 7.93DD2 pKa = 4.9KK3 pKa = 10.93KK4 pKa = 10.68DD5 pKa = 3.54GKK7 pKa = 9.07RR8 pKa = 11.84TRR10 pKa = 11.84TEE12 pKa = 3.84EE13 pKa = 4.0PPLRR17 pKa = 11.84EE18 pKa = 3.45VWRR21 pKa = 11.84QVMSEE26 pKa = 3.87YY27 pKa = 10.39RR28 pKa = 11.84SRR30 pKa = 11.84YY31 pKa = 7.31PGKK34 pKa = 9.53ATRR37 pKa = 11.84QGLWGMRR44 pKa = 11.84LVVIIGIMGQLSSGAEE60 pKa = 4.22YY61 pKa = 8.76ITLVSDD67 pKa = 4.24PYY69 pKa = 10.67GFKK72 pKa = 10.64PIRR75 pKa = 11.84DD76 pKa = 3.74VTNVPVTCVTKK87 pKa = 10.99NFTQWGCQPEE97 pKa = 4.79GAYY100 pKa = 9.72PDD102 pKa = 3.86PEE104 pKa = 3.94KK105 pKa = 10.55EE106 pKa = 3.81YY107 pKa = 11.08RR108 pKa = 11.84NISRR112 pKa = 11.84EE113 pKa = 3.87ILEE116 pKa = 4.08EE117 pKa = 4.37VYY119 pKa = 11.17NKK121 pKa = 7.9TWPWRR126 pKa = 11.84TYY128 pKa = 10.03HH129 pKa = 5.67WPLWQMEE136 pKa = 3.93NMRR139 pKa = 11.84DD140 pKa = 3.19WAEE143 pKa = 4.09QNLQNNKK150 pKa = 7.83TRR152 pKa = 11.84PEE154 pKa = 3.94IEE156 pKa = 4.0KK157 pKa = 10.72LLAGEE162 pKa = 3.73IRR164 pKa = 11.84GRR166 pKa = 11.84FCVPYY171 pKa = 9.08PYY173 pKa = 11.63AMIKK177 pKa = 10.1CEE179 pKa = 3.78EE180 pKa = 4.15WCWYY184 pKa = 8.75KK185 pKa = 11.11ARR187 pKa = 11.84GKK189 pKa = 9.89IQINCTRR196 pKa = 11.84AVVVSCTHH204 pKa = 5.61NMPLAGIKK212 pKa = 9.49RR213 pKa = 11.84VYY215 pKa = 9.0WDD217 pKa = 3.66KK218 pKa = 11.48KK219 pKa = 10.31DD220 pKa = 3.67EE221 pKa = 4.25KK222 pKa = 10.83EE223 pKa = 4.03MKK225 pKa = 10.26FMNIKK230 pKa = 10.12ACNTSAMRR238 pKa = 11.84CHH240 pKa = 6.28DD241 pKa = 3.69TKK243 pKa = 11.15VNPGGCVEE251 pKa = 5.37GYY253 pKa = 8.46PIPVGTEE260 pKa = 3.97IIPEE264 pKa = 4.09SFKK267 pKa = 11.04DD268 pKa = 3.5LRR270 pKa = 11.84EE271 pKa = 4.01EE272 pKa = 3.89KK273 pKa = 10.57SPYY276 pKa = 10.69GVIKK280 pKa = 10.6DD281 pKa = 3.75AAGKK285 pKa = 9.78LDD287 pKa = 3.28KK288 pKa = 10.63PLTVRR293 pKa = 11.84VWVRR297 pKa = 11.84LANVTGWVNGTPPFWQHH314 pKa = 7.02RR315 pKa = 11.84INGSTGINGTRR326 pKa = 11.84WYY328 pKa = 9.32GTEE331 pKa = 3.87TLTHH335 pKa = 6.63LGYY338 pKa = 10.77NISSNPEE345 pKa = 3.32AGICNKK351 pKa = 9.0TGKK354 pKa = 9.58IRR356 pKa = 11.84IGNKK360 pKa = 8.78EE361 pKa = 3.98FNLVYY366 pKa = 10.46APSWNCSQNWTGYY379 pKa = 8.04PVWHH383 pKa = 7.35IFRR386 pKa = 11.84HH387 pKa = 5.99LDD389 pKa = 3.19MTEE392 pKa = 4.37DD393 pKa = 3.24KK394 pKa = 10.6TSRR397 pKa = 11.84CMQRR401 pKa = 11.84PQRR404 pKa = 11.84KK405 pKa = 9.52NITIGNNTVIGNCSAQKK422 pKa = 10.02WDD424 pKa = 3.21GCTCNRR430 pKa = 11.84SGNWLRR436 pKa = 11.84NSTTGNIMIVVCRR449 pKa = 11.84EE450 pKa = 3.2NATLVGILEE459 pKa = 4.42GEE461 pKa = 4.45KK462 pKa = 10.34NWTQVDD468 pKa = 3.75EE469 pKa = 4.58IMRR472 pKa = 11.84KK473 pKa = 8.4NCSSEE478 pKa = 4.07EE479 pKa = 4.0QCNGTGIEE487 pKa = 4.02ALRR490 pKa = 11.84NIGGGTLGGVQNTNCTLPHH509 pKa = 6.45RR510 pKa = 11.84NEE512 pKa = 4.12TGHH515 pKa = 5.37WTCKK519 pKa = 9.86PRR521 pKa = 11.84TKK523 pKa = 10.33EE524 pKa = 3.81GKK526 pKa = 8.46TDD528 pKa = 3.11SLYY531 pKa = 10.4IGGKK535 pKa = 9.36KK536 pKa = 10.11LYY538 pKa = 9.94EE539 pKa = 3.96RR540 pKa = 11.84VKK542 pKa = 10.68AQYY545 pKa = 10.4SCEE548 pKa = 4.2SNLGGIDD555 pKa = 3.96VMVHH559 pKa = 4.37QQLMLQKK566 pKa = 8.69YY567 pKa = 9.39QIVQVRR573 pKa = 11.84TYY575 pKa = 8.76TYY577 pKa = 10.94GVVEE581 pKa = 4.71MPAEE585 pKa = 3.91YY586 pKa = 10.65AEE588 pKa = 4.1PSRR591 pKa = 11.84RR592 pKa = 11.84RR593 pKa = 11.84KK594 pKa = 10.07KK595 pKa = 10.48RR596 pKa = 11.84DD597 pKa = 2.83TSLARR602 pKa = 11.84KK603 pKa = 9.04KK604 pKa = 10.56RR605 pKa = 11.84GVGLVIVLVIMAIIAVAGASLGVANTVQQSEE636 pKa = 4.42TRR638 pKa = 11.84TAVQEE643 pKa = 4.14LANATAIQQQVLEE656 pKa = 4.24AAYY659 pKa = 10.91AMVQHH664 pKa = 5.89VAKK667 pKa = 10.52GIRR670 pKa = 11.84ILEE673 pKa = 3.97ARR675 pKa = 11.84VARR678 pKa = 11.84VEE680 pKa = 4.0ALVDD684 pKa = 3.88RR685 pKa = 11.84MMLYY689 pKa = 10.13QEE691 pKa = 4.12IDD693 pKa = 2.94CWHH696 pKa = 5.38YY697 pKa = 10.71HH698 pKa = 6.05GYY700 pKa = 10.41CVTSTRR706 pKa = 11.84SEE708 pKa = 3.94VANYY712 pKa = 10.0INWTRR717 pKa = 11.84YY718 pKa = 9.52RR719 pKa = 11.84DD720 pKa = 3.46NCTWQEE726 pKa = 3.75WEE728 pKa = 4.42KK729 pKa = 10.84EE730 pKa = 4.16IEE732 pKa = 4.0AHH734 pKa = 6.55EE735 pKa = 4.79GNLTQLIRR743 pKa = 11.84QSALATHH750 pKa = 7.09IAHH753 pKa = 7.4RR754 pKa = 11.84DD755 pKa = 3.09ASRR758 pKa = 11.84IPDD761 pKa = 3.16VWKK764 pKa = 10.56ALQQAFNWSGWFSWLKK780 pKa = 8.72YY781 pKa = 9.8LPWVIVGIIGIICIRR796 pKa = 11.84IGACVLGICAQSYY809 pKa = 9.46RR810 pKa = 11.84QLKK813 pKa = 8.66SLQYY817 pKa = 10.44QIIEE821 pKa = 3.82LDD823 pKa = 3.67LRR825 pKa = 11.84EE826 pKa = 4.07KK827 pKa = 10.91AIGCEE832 pKa = 4.23GEE834 pKa = 4.15EE835 pKa = 4.41GGEE838 pKa = 4.24DD839 pKa = 3.43EE840 pKa = 5.48AGSDD844 pKa = 3.6GFANCVLEE852 pKa = 4.91DD853 pKa = 4.18SPSLIQIWRR862 pKa = 11.84TQYY865 pKa = 10.22QEE867 pKa = 3.51WRR869 pKa = 11.84RR870 pKa = 11.84SPWATLEE877 pKa = 4.0KK878 pKa = 10.76ARR880 pKa = 11.84TLMVLPVTATEE891 pKa = 3.75QWLRR895 pKa = 11.84DD896 pKa = 3.72CGRR899 pKa = 11.84LGDD902 pKa = 3.78RR903 pKa = 11.84LKK905 pKa = 11.08NKK907 pKa = 9.71KK908 pKa = 9.86KK909 pKa = 10.55RR910 pKa = 11.84VDD912 pKa = 3.59CEE914 pKa = 4.21GQEE917 pKa = 4.34SRR919 pKa = 11.84PHH921 pKa = 4.48WTEE924 pKa = 3.47TT925 pKa = 3.27
MM1 pKa = 7.93DD2 pKa = 4.9KK3 pKa = 10.93KK4 pKa = 10.68DD5 pKa = 3.54GKK7 pKa = 9.07RR8 pKa = 11.84TRR10 pKa = 11.84TEE12 pKa = 3.84EE13 pKa = 4.0PPLRR17 pKa = 11.84EE18 pKa = 3.45VWRR21 pKa = 11.84QVMSEE26 pKa = 3.87YY27 pKa = 10.39RR28 pKa = 11.84SRR30 pKa = 11.84YY31 pKa = 7.31PGKK34 pKa = 9.53ATRR37 pKa = 11.84QGLWGMRR44 pKa = 11.84LVVIIGIMGQLSSGAEE60 pKa = 4.22YY61 pKa = 8.76ITLVSDD67 pKa = 4.24PYY69 pKa = 10.67GFKK72 pKa = 10.64PIRR75 pKa = 11.84DD76 pKa = 3.74VTNVPVTCVTKK87 pKa = 10.99NFTQWGCQPEE97 pKa = 4.79GAYY100 pKa = 9.72PDD102 pKa = 3.86PEE104 pKa = 3.94KK105 pKa = 10.55EE106 pKa = 3.81YY107 pKa = 11.08RR108 pKa = 11.84NISRR112 pKa = 11.84EE113 pKa = 3.87ILEE116 pKa = 4.08EE117 pKa = 4.37VYY119 pKa = 11.17NKK121 pKa = 7.9TWPWRR126 pKa = 11.84TYY128 pKa = 10.03HH129 pKa = 5.67WPLWQMEE136 pKa = 3.93NMRR139 pKa = 11.84DD140 pKa = 3.19WAEE143 pKa = 4.09QNLQNNKK150 pKa = 7.83TRR152 pKa = 11.84PEE154 pKa = 3.94IEE156 pKa = 4.0KK157 pKa = 10.72LLAGEE162 pKa = 3.73IRR164 pKa = 11.84GRR166 pKa = 11.84FCVPYY171 pKa = 9.08PYY173 pKa = 11.63AMIKK177 pKa = 10.1CEE179 pKa = 3.78EE180 pKa = 4.15WCWYY184 pKa = 8.75KK185 pKa = 11.11ARR187 pKa = 11.84GKK189 pKa = 9.89IQINCTRR196 pKa = 11.84AVVVSCTHH204 pKa = 5.61NMPLAGIKK212 pKa = 9.49RR213 pKa = 11.84VYY215 pKa = 9.0WDD217 pKa = 3.66KK218 pKa = 11.48KK219 pKa = 10.31DD220 pKa = 3.67EE221 pKa = 4.25KK222 pKa = 10.83EE223 pKa = 4.03MKK225 pKa = 10.26FMNIKK230 pKa = 10.12ACNTSAMRR238 pKa = 11.84CHH240 pKa = 6.28DD241 pKa = 3.69TKK243 pKa = 11.15VNPGGCVEE251 pKa = 5.37GYY253 pKa = 8.46PIPVGTEE260 pKa = 3.97IIPEE264 pKa = 4.09SFKK267 pKa = 11.04DD268 pKa = 3.5LRR270 pKa = 11.84EE271 pKa = 4.01EE272 pKa = 3.89KK273 pKa = 10.57SPYY276 pKa = 10.69GVIKK280 pKa = 10.6DD281 pKa = 3.75AAGKK285 pKa = 9.78LDD287 pKa = 3.28KK288 pKa = 10.63PLTVRR293 pKa = 11.84VWVRR297 pKa = 11.84LANVTGWVNGTPPFWQHH314 pKa = 7.02RR315 pKa = 11.84INGSTGINGTRR326 pKa = 11.84WYY328 pKa = 9.32GTEE331 pKa = 3.87TLTHH335 pKa = 6.63LGYY338 pKa = 10.77NISSNPEE345 pKa = 3.32AGICNKK351 pKa = 9.0TGKK354 pKa = 9.58IRR356 pKa = 11.84IGNKK360 pKa = 8.78EE361 pKa = 3.98FNLVYY366 pKa = 10.46APSWNCSQNWTGYY379 pKa = 8.04PVWHH383 pKa = 7.35IFRR386 pKa = 11.84HH387 pKa = 5.99LDD389 pKa = 3.19MTEE392 pKa = 4.37DD393 pKa = 3.24KK394 pKa = 10.6TSRR397 pKa = 11.84CMQRR401 pKa = 11.84PQRR404 pKa = 11.84KK405 pKa = 9.52NITIGNNTVIGNCSAQKK422 pKa = 10.02WDD424 pKa = 3.21GCTCNRR430 pKa = 11.84SGNWLRR436 pKa = 11.84NSTTGNIMIVVCRR449 pKa = 11.84EE450 pKa = 3.2NATLVGILEE459 pKa = 4.42GEE461 pKa = 4.45KK462 pKa = 10.34NWTQVDD468 pKa = 3.75EE469 pKa = 4.58IMRR472 pKa = 11.84KK473 pKa = 8.4NCSSEE478 pKa = 4.07EE479 pKa = 4.0QCNGTGIEE487 pKa = 4.02ALRR490 pKa = 11.84NIGGGTLGGVQNTNCTLPHH509 pKa = 6.45RR510 pKa = 11.84NEE512 pKa = 4.12TGHH515 pKa = 5.37WTCKK519 pKa = 9.86PRR521 pKa = 11.84TKK523 pKa = 10.33EE524 pKa = 3.81GKK526 pKa = 8.46TDD528 pKa = 3.11SLYY531 pKa = 10.4IGGKK535 pKa = 9.36KK536 pKa = 10.11LYY538 pKa = 9.94EE539 pKa = 3.96RR540 pKa = 11.84VKK542 pKa = 10.68AQYY545 pKa = 10.4SCEE548 pKa = 4.2SNLGGIDD555 pKa = 3.96VMVHH559 pKa = 4.37QQLMLQKK566 pKa = 8.69YY567 pKa = 9.39QIVQVRR573 pKa = 11.84TYY575 pKa = 8.76TYY577 pKa = 10.94GVVEE581 pKa = 4.71MPAEE585 pKa = 3.91YY586 pKa = 10.65AEE588 pKa = 4.1PSRR591 pKa = 11.84RR592 pKa = 11.84RR593 pKa = 11.84KK594 pKa = 10.07KK595 pKa = 10.48RR596 pKa = 11.84DD597 pKa = 2.83TSLARR602 pKa = 11.84KK603 pKa = 9.04KK604 pKa = 10.56RR605 pKa = 11.84GVGLVIVLVIMAIIAVAGASLGVANTVQQSEE636 pKa = 4.42TRR638 pKa = 11.84TAVQEE643 pKa = 4.14LANATAIQQQVLEE656 pKa = 4.24AAYY659 pKa = 10.91AMVQHH664 pKa = 5.89VAKK667 pKa = 10.52GIRR670 pKa = 11.84ILEE673 pKa = 3.97ARR675 pKa = 11.84VARR678 pKa = 11.84VEE680 pKa = 4.0ALVDD684 pKa = 3.88RR685 pKa = 11.84MMLYY689 pKa = 10.13QEE691 pKa = 4.12IDD693 pKa = 2.94CWHH696 pKa = 5.38YY697 pKa = 10.71HH698 pKa = 6.05GYY700 pKa = 10.41CVTSTRR706 pKa = 11.84SEE708 pKa = 3.94VANYY712 pKa = 10.0INWTRR717 pKa = 11.84YY718 pKa = 9.52RR719 pKa = 11.84DD720 pKa = 3.46NCTWQEE726 pKa = 3.75WEE728 pKa = 4.42KK729 pKa = 10.84EE730 pKa = 4.16IEE732 pKa = 4.0AHH734 pKa = 6.55EE735 pKa = 4.79GNLTQLIRR743 pKa = 11.84QSALATHH750 pKa = 7.09IAHH753 pKa = 7.4RR754 pKa = 11.84DD755 pKa = 3.09ASRR758 pKa = 11.84IPDD761 pKa = 3.16VWKK764 pKa = 10.56ALQQAFNWSGWFSWLKK780 pKa = 8.72YY781 pKa = 9.8LPWVIVGIIGIICIRR796 pKa = 11.84IGACVLGICAQSYY809 pKa = 9.46RR810 pKa = 11.84QLKK813 pKa = 8.66SLQYY817 pKa = 10.44QIIEE821 pKa = 3.82LDD823 pKa = 3.67LRR825 pKa = 11.84EE826 pKa = 4.07KK827 pKa = 10.91AIGCEE832 pKa = 4.23GEE834 pKa = 4.15EE835 pKa = 4.41GGEE838 pKa = 4.24DD839 pKa = 3.43EE840 pKa = 5.48AGSDD844 pKa = 3.6GFANCVLEE852 pKa = 4.91DD853 pKa = 4.18SPSLIQIWRR862 pKa = 11.84TQYY865 pKa = 10.22QEE867 pKa = 3.51WRR869 pKa = 11.84RR870 pKa = 11.84SPWATLEE877 pKa = 4.0KK878 pKa = 10.76ARR880 pKa = 11.84TLMVLPVTATEE891 pKa = 3.75QWLRR895 pKa = 11.84DD896 pKa = 3.72CGRR899 pKa = 11.84LGDD902 pKa = 3.78RR903 pKa = 11.84LKK905 pKa = 11.08NKK907 pKa = 9.71KK908 pKa = 9.86KK909 pKa = 10.55RR910 pKa = 11.84VDD912 pKa = 3.59CEE914 pKa = 4.21GQEE917 pKa = 4.34SRR919 pKa = 11.84PHH921 pKa = 4.48WTEE924 pKa = 3.47TT925 pKa = 3.27
Molecular weight: 105.86 kDa
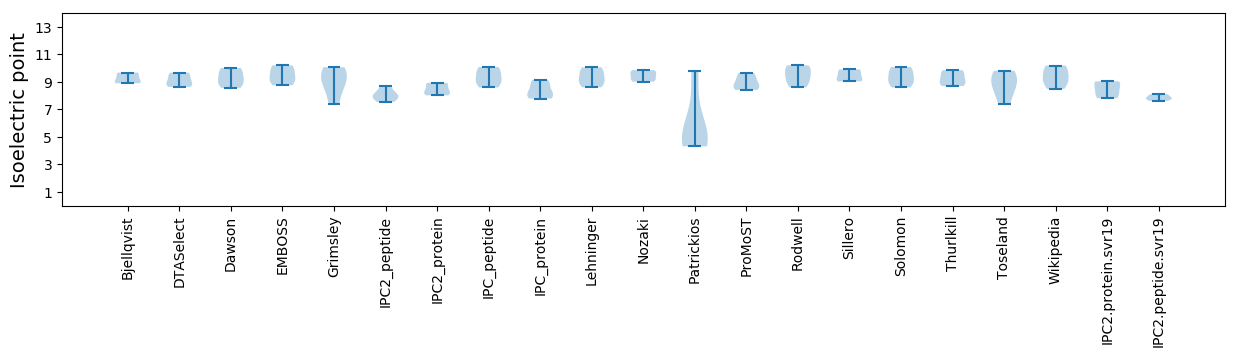
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C9DI74|C9DI74_CAEV Rev protein OS=Caprine arthritis encephalitis virus OX=11660 GN=rev PE=4 SV=1
MM1 pKa = 7.93DD2 pKa = 4.9KK3 pKa = 10.93KK4 pKa = 10.68DD5 pKa = 3.54GKK7 pKa = 9.07RR8 pKa = 11.84TRR10 pKa = 11.84TEE12 pKa = 3.84EE13 pKa = 4.0PPLRR17 pKa = 11.84EE18 pKa = 3.45VWRR21 pKa = 11.84QVMSEE26 pKa = 3.84YY27 pKa = 10.26RR28 pKa = 11.84SRR30 pKa = 11.84YY31 pKa = 9.11PEE33 pKa = 3.99LQATEE38 pKa = 4.09EE39 pKa = 4.34LAVSDD44 pKa = 4.6HH45 pKa = 5.79RR46 pKa = 11.84TGPKK50 pKa = 10.06GEE52 pKa = 4.27GYY54 pKa = 10.7RR55 pKa = 11.84MRR57 pKa = 11.84GRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84GRR65 pKa = 11.84GWFRR69 pKa = 11.84WLRR72 pKa = 11.84KK73 pKa = 9.27LRR75 pKa = 11.84ARR77 pKa = 11.84RR78 pKa = 11.84FTEE81 pKa = 4.08PDD83 pKa = 3.51PDD85 pKa = 4.72LEE87 pKa = 4.75DD88 pKa = 4.19PVPGMEE94 pKa = 4.38TLTLGDD100 pKa = 4.29PGEE103 pKa = 4.56GADD106 pKa = 3.74TDD108 pKa = 4.32GAAGDD113 pKa = 3.75SDD115 pKa = 4.03GAVAAGLWTAWRR127 pKa = 11.84SPQKK131 pKa = 10.92
MM1 pKa = 7.93DD2 pKa = 4.9KK3 pKa = 10.93KK4 pKa = 10.68DD5 pKa = 3.54GKK7 pKa = 9.07RR8 pKa = 11.84TRR10 pKa = 11.84TEE12 pKa = 3.84EE13 pKa = 4.0PPLRR17 pKa = 11.84EE18 pKa = 3.45VWRR21 pKa = 11.84QVMSEE26 pKa = 3.84YY27 pKa = 10.26RR28 pKa = 11.84SRR30 pKa = 11.84YY31 pKa = 9.11PEE33 pKa = 3.99LQATEE38 pKa = 4.09EE39 pKa = 4.34LAVSDD44 pKa = 4.6HH45 pKa = 5.79RR46 pKa = 11.84TGPKK50 pKa = 10.06GEE52 pKa = 4.27GYY54 pKa = 10.7RR55 pKa = 11.84MRR57 pKa = 11.84GRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84GRR65 pKa = 11.84GWFRR69 pKa = 11.84WLRR72 pKa = 11.84KK73 pKa = 9.27LRR75 pKa = 11.84ARR77 pKa = 11.84RR78 pKa = 11.84FTEE81 pKa = 4.08PDD83 pKa = 3.51PDD85 pKa = 4.72LEE87 pKa = 4.75DD88 pKa = 4.19PVPGMEE94 pKa = 4.38TLTLGDD100 pKa = 4.29PGEE103 pKa = 4.56GADD106 pKa = 3.74TDD108 pKa = 4.32GAAGDD113 pKa = 3.75SDD115 pKa = 4.03GAVAAGLWTAWRR127 pKa = 11.84SPQKK131 pKa = 10.92
Molecular weight: 15.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2706 |
131 |
981 |
541.2 |
62.14 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.098 ± 0.609 | 2.476 ± 0.676 |
4.028 ± 0.562 | 7.613 ± 0.487 |
1.959 ± 0.434 | 7.724 ± 0.416 |
2.217 ± 0.324 | 5.802 ± 1.187 |
7.28 ± 0.7 | 7.724 ± 0.616 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.624 ± 0.25 | 3.88 ± 0.71 |
5.1 ± 0.513 | 5.876 ± 0.671 |
7.058 ± 1.157 | 4.102 ± 0.203 |
5.802 ± 0.466 | 5.876 ± 0.486 |
3.474 ± 0.621 | 3.289 ± 0.296 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
