
Roseivirga ehrenbergii (strain DSM 102268 / JCM 13514 / KCTC 12282 / NCIMB 14502 / KMM 6017)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales;
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
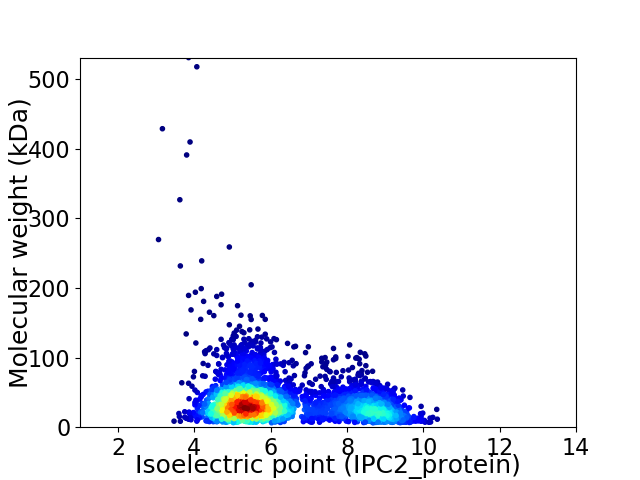
Virtual 2D-PAGE plot for 3632 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
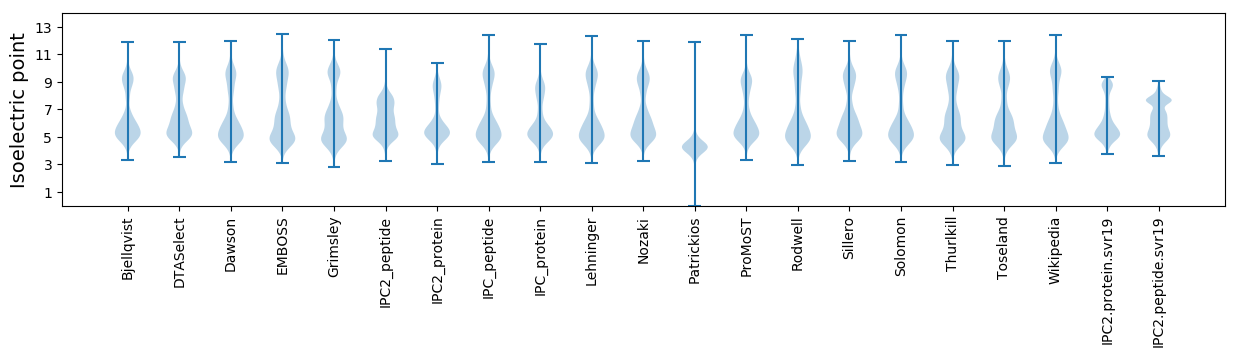
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A150XLH7|A0A150XLH7_ROSEK POTRA domain-containing protein OS=Roseivirga ehrenbergii (strain DSM 102268 / JCM 13514 / KCTC 12282 / NCIMB 14502 / KMM 6017) OX=279360 GN=MB14_17200 PE=4 SV=1
MM1 pKa = 6.9VRR3 pKa = 11.84KK4 pKa = 7.9TRR6 pKa = 11.84ILLLLLGITFSVLTQAQNFTRR27 pKa = 11.84HH28 pKa = 4.91NWYY31 pKa = 10.37FSNNDD36 pKa = 2.85QALIFGKK43 pKa = 9.75EE44 pKa = 3.39QAARR48 pKa = 11.84SFIADD53 pKa = 3.49GKK55 pKa = 9.93IAQTNAGEE63 pKa = 4.52KK64 pKa = 9.13LTATDD69 pKa = 3.89PTTGDD74 pKa = 3.67LLFYY78 pKa = 10.88SDD80 pKa = 5.7GINIYY85 pKa = 10.31DD86 pKa = 3.54ATHH89 pKa = 7.03AIMEE93 pKa = 4.74NGTGLTTDD101 pKa = 3.85PNGIQPLALSPVPGVGNEE119 pKa = 3.23SRR121 pKa = 11.84YY122 pKa = 9.77YY123 pKa = 9.27VFHH126 pKa = 7.11RR127 pKa = 11.84NVTGEE132 pKa = 3.79ILYY135 pKa = 9.12TLVDD139 pKa = 3.71MSSQGNRR146 pKa = 11.84VDD148 pKa = 4.13GPPAGAVVIGSKK160 pKa = 9.42NQPTGIISRR169 pKa = 11.84GDD171 pKa = 3.16GMLTVASGDD180 pKa = 3.41MTQFWLVTQNSTNGNFEE197 pKa = 3.59IHH199 pKa = 7.25EE200 pKa = 4.43IPVAGQPFDD209 pKa = 3.75PQGAPTVLTANVQAAHH225 pKa = 7.02LSLNPMGTQIAVRR238 pKa = 11.84PSNNVNIQILDD249 pKa = 3.71FNRR252 pKa = 11.84DD253 pKa = 3.28TPEE256 pKa = 3.78LMFNRR261 pKa = 11.84SVANSFVPNQTLTGSIGWSNDD282 pKa = 2.38GTKK285 pKa = 10.48LYY287 pKa = 10.67FSRR290 pKa = 11.84NLANSTSGNLYY301 pKa = 10.41RR302 pKa = 11.84IDD304 pKa = 4.88LSDD307 pKa = 3.41PMASVEE313 pKa = 4.32PVFSNPIGASQSVMLAPDD331 pKa = 3.03SSLYY335 pKa = 10.51HH336 pKa = 6.62IYY338 pKa = 10.61KK339 pKa = 8.39EE340 pKa = 4.29TAAGPRR346 pKa = 11.84LLARR350 pKa = 11.84INQPDD355 pKa = 3.72SALTALDD362 pKa = 4.17YY363 pKa = 11.13EE364 pKa = 4.57PSLFDD369 pKa = 3.82EE370 pKa = 5.39FDD372 pKa = 3.59PASDD376 pKa = 3.81YY377 pKa = 10.59FQQFMPPAAIGPTVTISDD395 pKa = 4.07DD396 pKa = 3.47GDD398 pKa = 3.68YY399 pKa = 11.58CMNTPVQFYY408 pKa = 11.34VNIDD412 pKa = 3.55PSTAIPTSYY421 pKa = 10.51SWDD424 pKa = 3.33IQPFGITSNAIAPIFTFDD442 pKa = 3.24QAGMYY447 pKa = 7.39MATLTVEE454 pKa = 4.25FDD456 pKa = 4.19GIGTVMSNTLMGEE469 pKa = 3.51IMEE472 pKa = 4.83NDD474 pKa = 3.31IQVSLQDD481 pKa = 3.37TTICEE486 pKa = 4.58GEE488 pKa = 4.5TLTLDD493 pKa = 4.69AEE495 pKa = 4.44PQSGGQGQQGGATGGPFEE513 pKa = 4.4YY514 pKa = 9.96RR515 pKa = 11.84WNTGEE520 pKa = 4.18TTSSIDD526 pKa = 3.17VTDD529 pKa = 4.11AGNYY533 pKa = 6.32WVVITPTTGCPIYY546 pKa = 9.66EE547 pKa = 4.17TVEE550 pKa = 3.94VKK552 pKa = 10.8VYY554 pKa = 11.03GDD556 pKa = 3.99EE557 pKa = 4.44NPTANIWYY565 pKa = 8.85FGNGAGIDD573 pKa = 3.89FNEE576 pKa = 4.31VDD578 pKa = 4.56GLAPPPRR585 pKa = 11.84SITTAHH591 pKa = 6.83AMNAPEE597 pKa = 4.36GTSTISDD604 pKa = 3.51ANGQVLFYY612 pKa = 11.07TNGTSVWNADD622 pKa = 3.4NGVMPNGTKK631 pKa = 9.86IGGDD635 pKa = 3.57STSTQAVVIVPFEE648 pKa = 4.52DD649 pKa = 5.32DD650 pKa = 3.02EE651 pKa = 4.68TLFYY655 pKa = 11.31VFTTEE660 pKa = 4.73EE661 pKa = 3.92VHH663 pKa = 7.41GDD665 pKa = 3.14NTYY668 pKa = 8.38MLKK671 pKa = 10.75YY672 pKa = 10.47SVVDD676 pKa = 4.09LKK678 pKa = 11.23EE679 pKa = 4.44DD680 pKa = 3.44NGLGAVISKK689 pKa = 10.96DD690 pKa = 3.06NVLFTRR696 pKa = 11.84STEE699 pKa = 3.84KK700 pKa = 10.98LIALEE705 pKa = 4.14GAGGYY710 pKa = 8.97WLMAHH715 pKa = 7.36EE716 pKa = 4.92YY717 pKa = 8.48GTNTFRR723 pKa = 11.84AYY725 pKa = 9.96PITNQGIGQPILSSVGSVHH744 pKa = 7.26SINDD748 pKa = 3.7ALSGQAGMKK757 pKa = 10.41FSGDD761 pKa = 3.23RR762 pKa = 11.84LRR764 pKa = 11.84IAVALIEE771 pKa = 4.38GNEE774 pKa = 4.23DD775 pKa = 3.53YY776 pKa = 11.62VEE778 pKa = 4.45LFSFDD783 pKa = 4.07ANTGEE788 pKa = 4.41VTDD791 pKa = 5.08LLYY794 pKa = 10.89QIDD797 pKa = 4.28LGEE800 pKa = 4.83GGPALNDD807 pKa = 3.12QVYY810 pKa = 10.2DD811 pKa = 3.43VHH813 pKa = 7.54FSTGGDD819 pKa = 3.17KK820 pKa = 11.09LFATMNNRR828 pKa = 11.84NGGAPGGRR836 pKa = 11.84ILEE839 pKa = 3.97YY840 pKa = 10.44RR841 pKa = 11.84IDD843 pKa = 3.78SASTEE848 pKa = 4.25VTRR851 pKa = 11.84LASKK855 pKa = 9.88TDD857 pKa = 2.92ISTSSTVTVNYY868 pKa = 10.34GAIQTGPDD876 pKa = 3.31GQLYY880 pKa = 9.53VAMEE884 pKa = 4.19IPGNPAGTPFIGSINVNEE902 pKa = 4.1DD903 pKa = 3.04TAAVSSFNQMQVPLTTGNSRR923 pKa = 11.84LGLPNFVQNNVNPPMEE939 pKa = 4.93PSMSAPDD946 pKa = 3.64STCVEE951 pKa = 4.04EE952 pKa = 5.47RR953 pKa = 11.84IEE955 pKa = 3.92LSGVGTSDD963 pKa = 2.73IDD965 pKa = 3.54EE966 pKa = 4.6FLWTIIDD973 pKa = 3.99DD974 pKa = 4.22ANTTVFSASGQDD986 pKa = 3.17TAYY989 pKa = 10.8VFPQGQAGRR998 pKa = 11.84FDD1000 pKa = 3.17ISLNISNRR1008 pKa = 11.84CGFDD1012 pKa = 2.92TTFVQTIDD1020 pKa = 3.5VFEE1023 pKa = 4.43IPPRR1027 pKa = 11.84PAVPTAVVLCEE1038 pKa = 4.31GSSHH1042 pKa = 7.47DD1043 pKa = 4.44LDD1045 pKa = 4.52ALGGEE1050 pKa = 4.59PDD1052 pKa = 4.56DD1053 pKa = 4.27PTLSFEE1059 pKa = 4.16WTNSQGAVVSNSRR1072 pKa = 11.84YY1073 pKa = 8.85YY1074 pKa = 10.93SVTEE1078 pKa = 3.8EE1079 pKa = 4.26EE1080 pKa = 5.27IYY1082 pKa = 10.6TITITSVGGGGCVSSAEE1099 pKa = 4.35VYY1101 pKa = 9.81AAPPFEE1107 pKa = 5.63IEE1109 pKa = 4.59LPEE1112 pKa = 4.05TANLCEE1118 pKa = 4.55GEE1120 pKa = 4.66SLTLDD1125 pKa = 3.54PNVNANNYY1133 pKa = 7.51IWTRR1137 pKa = 11.84INPDD1141 pKa = 2.82NSTTTLPNQRR1151 pKa = 11.84TATVDD1156 pKa = 3.17SSVPGIFTYY1165 pKa = 10.59VVSIEE1170 pKa = 4.52DD1171 pKa = 4.86PIEE1174 pKa = 4.39PGCFANDD1181 pKa = 3.49STVVTINGSPNLSLVSSQDD1200 pKa = 3.04ASCPQVGPTPDD1211 pKa = 3.34GEE1213 pKa = 4.64FVFNIAGTGNYY1224 pKa = 9.98SYY1226 pKa = 10.07TVSNATGATVAQSNSVNGPNDD1247 pKa = 3.25VSVTGLDD1254 pKa = 3.33GGIYY1258 pKa = 9.9SVNVTDD1264 pKa = 5.45NSTNCTDD1271 pKa = 3.44EE1272 pKa = 3.99LTGIEE1277 pKa = 3.89IQNNNFNIIEE1287 pKa = 4.24PVVTADD1293 pKa = 3.79ATCDD1297 pKa = 3.42TADD1300 pKa = 3.4GSITITLDD1308 pKa = 3.4DD1309 pKa = 4.29ASVFPVSYY1317 pKa = 11.04VLTRR1321 pKa = 11.84DD1322 pKa = 3.36EE1323 pKa = 5.54AGFTPLSGNAVANGFVISGLEE1344 pKa = 3.8GGTYY1348 pKa = 10.08SIEE1351 pKa = 4.23VTSTALGCTATASGIVVNQPTDD1373 pKa = 3.47VQNLQISGTTEE1384 pKa = 3.57VCDD1387 pKa = 3.69ISPSTTLTATSNNGTNYY1404 pKa = 8.87TWIGPSGPAGNTASITVTEE1423 pKa = 4.33SGTYY1427 pKa = 8.94TVTADD1432 pKa = 3.81APGVACPTSEE1442 pKa = 4.26SVIVTLTVQPDD1453 pKa = 3.45VEE1455 pKa = 4.82IIQIGDD1461 pKa = 3.5VCDD1464 pKa = 3.66GTLVLEE1470 pKa = 4.17AQINNEE1476 pKa = 4.26NPTATYY1482 pKa = 10.72NFAWSTGEE1490 pKa = 3.77NTRR1493 pKa = 11.84SITVSADD1500 pKa = 2.66GTYY1503 pKa = 11.02DD1504 pKa = 3.28VTVRR1508 pKa = 11.84EE1509 pKa = 4.77AGDD1512 pKa = 3.62LTCTGTYY1519 pKa = 10.12SEE1521 pKa = 5.13TIDD1524 pKa = 3.89FPEE1527 pKa = 4.28EE1528 pKa = 3.86LEE1530 pKa = 4.27ATLSSSPACDD1540 pKa = 3.71DD1541 pKa = 3.84GQPITLTVDD1550 pKa = 3.5VLSGNATNFTWTRR1563 pKa = 11.84NGQAAGTGNPLVVNEE1578 pKa = 4.22EE1579 pKa = 3.59GSYY1582 pKa = 9.66TVTISDD1588 pKa = 3.6NSGCSIEE1595 pKa = 4.43RR1596 pKa = 11.84SILIRR1601 pKa = 11.84RR1602 pKa = 11.84SVIPEE1607 pKa = 3.85GLLPDD1612 pKa = 3.97LEE1614 pKa = 5.22YY1615 pKa = 11.46YY1616 pKa = 10.83CSTSSNVLTVTAGIGFEE1633 pKa = 4.29TYY1635 pKa = 8.99EE1636 pKa = 4.15WTLDD1640 pKa = 3.86GQPLAHH1646 pKa = 7.06SDD1648 pKa = 3.36QDD1650 pKa = 4.04LVIAAPGIYY1659 pKa = 9.26QVTMTTATGCVRR1671 pKa = 11.84VDD1673 pKa = 3.11QMEE1676 pKa = 4.73IIEE1679 pKa = 4.46SCDD1682 pKa = 3.52PVILAPNVIVPEE1694 pKa = 4.3GNPPNNVFSVVPNGFVSDD1712 pKa = 3.81FQIFIYY1718 pKa = 10.35SRR1720 pKa = 11.84WGEE1723 pKa = 4.31LIYY1726 pKa = 10.62QSSALEE1732 pKa = 4.09FQWNGTFNGQLVPIGTYY1749 pKa = 9.6PYY1751 pKa = 10.01VIKK1754 pKa = 10.5FRR1756 pKa = 11.84SRR1758 pKa = 11.84FQPEE1762 pKa = 3.49RR1763 pKa = 11.84GEE1765 pKa = 4.07FEE1767 pKa = 3.74QRR1769 pKa = 11.84GAITVVRR1776 pKa = 4.35
MM1 pKa = 6.9VRR3 pKa = 11.84KK4 pKa = 7.9TRR6 pKa = 11.84ILLLLLGITFSVLTQAQNFTRR27 pKa = 11.84HH28 pKa = 4.91NWYY31 pKa = 10.37FSNNDD36 pKa = 2.85QALIFGKK43 pKa = 9.75EE44 pKa = 3.39QAARR48 pKa = 11.84SFIADD53 pKa = 3.49GKK55 pKa = 9.93IAQTNAGEE63 pKa = 4.52KK64 pKa = 9.13LTATDD69 pKa = 3.89PTTGDD74 pKa = 3.67LLFYY78 pKa = 10.88SDD80 pKa = 5.7GINIYY85 pKa = 10.31DD86 pKa = 3.54ATHH89 pKa = 7.03AIMEE93 pKa = 4.74NGTGLTTDD101 pKa = 3.85PNGIQPLALSPVPGVGNEE119 pKa = 3.23SRR121 pKa = 11.84YY122 pKa = 9.77YY123 pKa = 9.27VFHH126 pKa = 7.11RR127 pKa = 11.84NVTGEE132 pKa = 3.79ILYY135 pKa = 9.12TLVDD139 pKa = 3.71MSSQGNRR146 pKa = 11.84VDD148 pKa = 4.13GPPAGAVVIGSKK160 pKa = 9.42NQPTGIISRR169 pKa = 11.84GDD171 pKa = 3.16GMLTVASGDD180 pKa = 3.41MTQFWLVTQNSTNGNFEE197 pKa = 3.59IHH199 pKa = 7.25EE200 pKa = 4.43IPVAGQPFDD209 pKa = 3.75PQGAPTVLTANVQAAHH225 pKa = 7.02LSLNPMGTQIAVRR238 pKa = 11.84PSNNVNIQILDD249 pKa = 3.71FNRR252 pKa = 11.84DD253 pKa = 3.28TPEE256 pKa = 3.78LMFNRR261 pKa = 11.84SVANSFVPNQTLTGSIGWSNDD282 pKa = 2.38GTKK285 pKa = 10.48LYY287 pKa = 10.67FSRR290 pKa = 11.84NLANSTSGNLYY301 pKa = 10.41RR302 pKa = 11.84IDD304 pKa = 4.88LSDD307 pKa = 3.41PMASVEE313 pKa = 4.32PVFSNPIGASQSVMLAPDD331 pKa = 3.03SSLYY335 pKa = 10.51HH336 pKa = 6.62IYY338 pKa = 10.61KK339 pKa = 8.39EE340 pKa = 4.29TAAGPRR346 pKa = 11.84LLARR350 pKa = 11.84INQPDD355 pKa = 3.72SALTALDD362 pKa = 4.17YY363 pKa = 11.13EE364 pKa = 4.57PSLFDD369 pKa = 3.82EE370 pKa = 5.39FDD372 pKa = 3.59PASDD376 pKa = 3.81YY377 pKa = 10.59FQQFMPPAAIGPTVTISDD395 pKa = 4.07DD396 pKa = 3.47GDD398 pKa = 3.68YY399 pKa = 11.58CMNTPVQFYY408 pKa = 11.34VNIDD412 pKa = 3.55PSTAIPTSYY421 pKa = 10.51SWDD424 pKa = 3.33IQPFGITSNAIAPIFTFDD442 pKa = 3.24QAGMYY447 pKa = 7.39MATLTVEE454 pKa = 4.25FDD456 pKa = 4.19GIGTVMSNTLMGEE469 pKa = 3.51IMEE472 pKa = 4.83NDD474 pKa = 3.31IQVSLQDD481 pKa = 3.37TTICEE486 pKa = 4.58GEE488 pKa = 4.5TLTLDD493 pKa = 4.69AEE495 pKa = 4.44PQSGGQGQQGGATGGPFEE513 pKa = 4.4YY514 pKa = 9.96RR515 pKa = 11.84WNTGEE520 pKa = 4.18TTSSIDD526 pKa = 3.17VTDD529 pKa = 4.11AGNYY533 pKa = 6.32WVVITPTTGCPIYY546 pKa = 9.66EE547 pKa = 4.17TVEE550 pKa = 3.94VKK552 pKa = 10.8VYY554 pKa = 11.03GDD556 pKa = 3.99EE557 pKa = 4.44NPTANIWYY565 pKa = 8.85FGNGAGIDD573 pKa = 3.89FNEE576 pKa = 4.31VDD578 pKa = 4.56GLAPPPRR585 pKa = 11.84SITTAHH591 pKa = 6.83AMNAPEE597 pKa = 4.36GTSTISDD604 pKa = 3.51ANGQVLFYY612 pKa = 11.07TNGTSVWNADD622 pKa = 3.4NGVMPNGTKK631 pKa = 9.86IGGDD635 pKa = 3.57STSTQAVVIVPFEE648 pKa = 4.52DD649 pKa = 5.32DD650 pKa = 3.02EE651 pKa = 4.68TLFYY655 pKa = 11.31VFTTEE660 pKa = 4.73EE661 pKa = 3.92VHH663 pKa = 7.41GDD665 pKa = 3.14NTYY668 pKa = 8.38MLKK671 pKa = 10.75YY672 pKa = 10.47SVVDD676 pKa = 4.09LKK678 pKa = 11.23EE679 pKa = 4.44DD680 pKa = 3.44NGLGAVISKK689 pKa = 10.96DD690 pKa = 3.06NVLFTRR696 pKa = 11.84STEE699 pKa = 3.84KK700 pKa = 10.98LIALEE705 pKa = 4.14GAGGYY710 pKa = 8.97WLMAHH715 pKa = 7.36EE716 pKa = 4.92YY717 pKa = 8.48GTNTFRR723 pKa = 11.84AYY725 pKa = 9.96PITNQGIGQPILSSVGSVHH744 pKa = 7.26SINDD748 pKa = 3.7ALSGQAGMKK757 pKa = 10.41FSGDD761 pKa = 3.23RR762 pKa = 11.84LRR764 pKa = 11.84IAVALIEE771 pKa = 4.38GNEE774 pKa = 4.23DD775 pKa = 3.53YY776 pKa = 11.62VEE778 pKa = 4.45LFSFDD783 pKa = 4.07ANTGEE788 pKa = 4.41VTDD791 pKa = 5.08LLYY794 pKa = 10.89QIDD797 pKa = 4.28LGEE800 pKa = 4.83GGPALNDD807 pKa = 3.12QVYY810 pKa = 10.2DD811 pKa = 3.43VHH813 pKa = 7.54FSTGGDD819 pKa = 3.17KK820 pKa = 11.09LFATMNNRR828 pKa = 11.84NGGAPGGRR836 pKa = 11.84ILEE839 pKa = 3.97YY840 pKa = 10.44RR841 pKa = 11.84IDD843 pKa = 3.78SASTEE848 pKa = 4.25VTRR851 pKa = 11.84LASKK855 pKa = 9.88TDD857 pKa = 2.92ISTSSTVTVNYY868 pKa = 10.34GAIQTGPDD876 pKa = 3.31GQLYY880 pKa = 9.53VAMEE884 pKa = 4.19IPGNPAGTPFIGSINVNEE902 pKa = 4.1DD903 pKa = 3.04TAAVSSFNQMQVPLTTGNSRR923 pKa = 11.84LGLPNFVQNNVNPPMEE939 pKa = 4.93PSMSAPDD946 pKa = 3.64STCVEE951 pKa = 4.04EE952 pKa = 5.47RR953 pKa = 11.84IEE955 pKa = 3.92LSGVGTSDD963 pKa = 2.73IDD965 pKa = 3.54EE966 pKa = 4.6FLWTIIDD973 pKa = 3.99DD974 pKa = 4.22ANTTVFSASGQDD986 pKa = 3.17TAYY989 pKa = 10.8VFPQGQAGRR998 pKa = 11.84FDD1000 pKa = 3.17ISLNISNRR1008 pKa = 11.84CGFDD1012 pKa = 2.92TTFVQTIDD1020 pKa = 3.5VFEE1023 pKa = 4.43IPPRR1027 pKa = 11.84PAVPTAVVLCEE1038 pKa = 4.31GSSHH1042 pKa = 7.47DD1043 pKa = 4.44LDD1045 pKa = 4.52ALGGEE1050 pKa = 4.59PDD1052 pKa = 4.56DD1053 pKa = 4.27PTLSFEE1059 pKa = 4.16WTNSQGAVVSNSRR1072 pKa = 11.84YY1073 pKa = 8.85YY1074 pKa = 10.93SVTEE1078 pKa = 3.8EE1079 pKa = 4.26EE1080 pKa = 5.27IYY1082 pKa = 10.6TITITSVGGGGCVSSAEE1099 pKa = 4.35VYY1101 pKa = 9.81AAPPFEE1107 pKa = 5.63IEE1109 pKa = 4.59LPEE1112 pKa = 4.05TANLCEE1118 pKa = 4.55GEE1120 pKa = 4.66SLTLDD1125 pKa = 3.54PNVNANNYY1133 pKa = 7.51IWTRR1137 pKa = 11.84INPDD1141 pKa = 2.82NSTTTLPNQRR1151 pKa = 11.84TATVDD1156 pKa = 3.17SSVPGIFTYY1165 pKa = 10.59VVSIEE1170 pKa = 4.52DD1171 pKa = 4.86PIEE1174 pKa = 4.39PGCFANDD1181 pKa = 3.49STVVTINGSPNLSLVSSQDD1200 pKa = 3.04ASCPQVGPTPDD1211 pKa = 3.34GEE1213 pKa = 4.64FVFNIAGTGNYY1224 pKa = 9.98SYY1226 pKa = 10.07TVSNATGATVAQSNSVNGPNDD1247 pKa = 3.25VSVTGLDD1254 pKa = 3.33GGIYY1258 pKa = 9.9SVNVTDD1264 pKa = 5.45NSTNCTDD1271 pKa = 3.44EE1272 pKa = 3.99LTGIEE1277 pKa = 3.89IQNNNFNIIEE1287 pKa = 4.24PVVTADD1293 pKa = 3.79ATCDD1297 pKa = 3.42TADD1300 pKa = 3.4GSITITLDD1308 pKa = 3.4DD1309 pKa = 4.29ASVFPVSYY1317 pKa = 11.04VLTRR1321 pKa = 11.84DD1322 pKa = 3.36EE1323 pKa = 5.54AGFTPLSGNAVANGFVISGLEE1344 pKa = 3.8GGTYY1348 pKa = 10.08SIEE1351 pKa = 4.23VTSTALGCTATASGIVVNQPTDD1373 pKa = 3.47VQNLQISGTTEE1384 pKa = 3.57VCDD1387 pKa = 3.69ISPSTTLTATSNNGTNYY1404 pKa = 8.87TWIGPSGPAGNTASITVTEE1423 pKa = 4.33SGTYY1427 pKa = 8.94TVTADD1432 pKa = 3.81APGVACPTSEE1442 pKa = 4.26SVIVTLTVQPDD1453 pKa = 3.45VEE1455 pKa = 4.82IIQIGDD1461 pKa = 3.5VCDD1464 pKa = 3.66GTLVLEE1470 pKa = 4.17AQINNEE1476 pKa = 4.26NPTATYY1482 pKa = 10.72NFAWSTGEE1490 pKa = 3.77NTRR1493 pKa = 11.84SITVSADD1500 pKa = 2.66GTYY1503 pKa = 11.02DD1504 pKa = 3.28VTVRR1508 pKa = 11.84EE1509 pKa = 4.77AGDD1512 pKa = 3.62LTCTGTYY1519 pKa = 10.12SEE1521 pKa = 5.13TIDD1524 pKa = 3.89FPEE1527 pKa = 4.28EE1528 pKa = 3.86LEE1530 pKa = 4.27ATLSSSPACDD1540 pKa = 3.71DD1541 pKa = 3.84GQPITLTVDD1550 pKa = 3.5VLSGNATNFTWTRR1563 pKa = 11.84NGQAAGTGNPLVVNEE1578 pKa = 4.22EE1579 pKa = 3.59GSYY1582 pKa = 9.66TVTISDD1588 pKa = 3.6NSGCSIEE1595 pKa = 4.43RR1596 pKa = 11.84SILIRR1601 pKa = 11.84RR1602 pKa = 11.84SVIPEE1607 pKa = 3.85GLLPDD1612 pKa = 3.97LEE1614 pKa = 5.22YY1615 pKa = 11.46YY1616 pKa = 10.83CSTSSNVLTVTAGIGFEE1633 pKa = 4.29TYY1635 pKa = 8.99EE1636 pKa = 4.15WTLDD1640 pKa = 3.86GQPLAHH1646 pKa = 7.06SDD1648 pKa = 3.36QDD1650 pKa = 4.04LVIAAPGIYY1659 pKa = 9.26QVTMTTATGCVRR1671 pKa = 11.84VDD1673 pKa = 3.11QMEE1676 pKa = 4.73IIEE1679 pKa = 4.46SCDD1682 pKa = 3.52PVILAPNVIVPEE1694 pKa = 4.3GNPPNNVFSVVPNGFVSDD1712 pKa = 3.81FQIFIYY1718 pKa = 10.35SRR1720 pKa = 11.84WGEE1723 pKa = 4.31LIYY1726 pKa = 10.62QSSALEE1732 pKa = 4.09FQWNGTFNGQLVPIGTYY1749 pKa = 9.6PYY1751 pKa = 10.01VIKK1754 pKa = 10.5FRR1756 pKa = 11.84SRR1758 pKa = 11.84FQPEE1762 pKa = 3.49RR1763 pKa = 11.84GEE1765 pKa = 4.07FEE1767 pKa = 3.74QRR1769 pKa = 11.84GAITVVRR1776 pKa = 4.35
Molecular weight: 189.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A150X0B7|A0A150X0B7_ROSEK Peptidase OS=Roseivirga ehrenbergii (strain DSM 102268 / JCM 13514 / KCTC 12282 / NCIMB 14502 / KMM 6017) OX=279360 GN=MB14_08900 PE=4 SV=1
MM1 pKa = 7.38TFLFEE6 pKa = 5.33LYY8 pKa = 9.78IKK10 pKa = 10.57RR11 pKa = 11.84LLGKK15 pKa = 9.38YY16 pKa = 9.33RR17 pKa = 11.84YY18 pKa = 9.24RR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 10.27SLLWHH26 pKa = 6.81LSRR29 pKa = 11.84GVNCGKK35 pKa = 9.56PLKK38 pKa = 10.4RR39 pKa = 11.84LMEE42 pKa = 4.32TLCSIAHH49 pKa = 6.1GLNRR53 pKa = 11.84GLFEE57 pKa = 6.39CITLGNGFNHH67 pKa = 6.29FNWVVGLLILPQRR80 pKa = 11.84ALSSAKK86 pKa = 10.09GKK88 pKa = 10.01LVQRR92 pKa = 11.84QQ93 pKa = 3.09
MM1 pKa = 7.38TFLFEE6 pKa = 5.33LYY8 pKa = 9.78IKK10 pKa = 10.57RR11 pKa = 11.84LLGKK15 pKa = 9.38YY16 pKa = 9.33RR17 pKa = 11.84YY18 pKa = 9.24RR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 10.27SLLWHH26 pKa = 6.81LSRR29 pKa = 11.84GVNCGKK35 pKa = 9.56PLKK38 pKa = 10.4RR39 pKa = 11.84LMEE42 pKa = 4.32TLCSIAHH49 pKa = 6.1GLNRR53 pKa = 11.84GLFEE57 pKa = 6.39CITLGNGFNHH67 pKa = 6.29FNWVVGLLILPQRR80 pKa = 11.84ALSSAKK86 pKa = 10.09GKK88 pKa = 10.01LVQRR92 pKa = 11.84QQ93 pKa = 3.09
Molecular weight: 10.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1259309 |
54 |
4957 |
346.7 |
38.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.802 ± 0.037 | 0.642 ± 0.012 |
5.576 ± 0.048 | 6.917 ± 0.05 |
5.185 ± 0.035 | 6.809 ± 0.047 |
1.713 ± 0.021 | 7.402 ± 0.032 |
7.01 ± 0.056 | 9.483 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.412 ± 0.025 | 5.524 ± 0.037 |
3.502 ± 0.023 | 3.509 ± 0.025 |
3.847 ± 0.029 | 6.737 ± 0.042 |
5.57 ± 0.061 | 6.46 ± 0.029 |
1.091 ± 0.017 | 3.809 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
