
Kwoniella mangroviensis CBS 10435
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Tremellomycetes; Tremellales; Cryptococcaceae; Kwoniella; Kwoniella mangrovensis
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
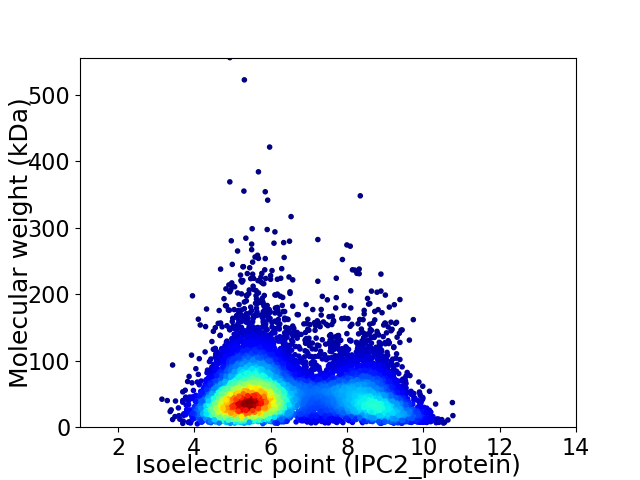
Virtual 2D-PAGE plot for 8428 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B9IP46|A0A1B9IP46_9TREE Elongator complex protein 5 OS=Kwoniella mangroviensis CBS 10435 OX=1331196 GN=L486_04804 PE=3 SV=1
MM1 pKa = 7.05QLSIIPFLTILSILFPTSSAQLDD24 pKa = 3.8TDD26 pKa = 4.52IICPDD31 pKa = 3.4GSLTEE36 pKa = 4.4YY37 pKa = 8.58LTSMMDD43 pKa = 3.25VLFANGLTTFEE54 pKa = 3.89QLIVHH59 pKa = 6.61WSEE62 pKa = 3.22TDD64 pKa = 2.81MGYY67 pKa = 10.75EE68 pKa = 4.14FLHH71 pKa = 6.74DD72 pKa = 5.3LYY74 pKa = 11.73NSGQKK79 pKa = 8.99LTVLVPTNDD88 pKa = 3.19AFQEE92 pKa = 4.19SGIVSPFEE100 pKa = 4.18GLTEE104 pKa = 4.05DD105 pKa = 3.22WGAEE109 pKa = 3.88LGEE112 pKa = 4.01LHH114 pKa = 7.25LLQGEE119 pKa = 4.25WTYY122 pKa = 11.83DD123 pKa = 3.34QIPQSGHH130 pKa = 5.49AVAATSLLLANEE142 pKa = 4.47LNSTDD147 pKa = 3.58IQPNAYY153 pKa = 9.27QAMVLEE159 pKa = 4.82RR160 pKa = 11.84GNDD163 pKa = 3.42DD164 pKa = 3.61SVVVNGWWGHH174 pKa = 4.69ATSTSGPLDD183 pKa = 3.59LSGSGGLLDD192 pKa = 4.75NLFILPIDD200 pKa = 3.73QVLSFPPSLSTALQAPGLTNMSSALQVIGKK230 pKa = 9.76SSDD233 pKa = 3.43VEE235 pKa = 4.1RR236 pKa = 11.84LSEE239 pKa = 4.11GGFTIFVPLDD249 pKa = 3.72SVWNDD254 pKa = 3.21EE255 pKa = 5.0LKK257 pKa = 10.88DD258 pKa = 3.57IMLDD262 pKa = 3.49GSKK265 pKa = 10.54APEE268 pKa = 3.87MIGNHH273 pKa = 5.15FTTSYY278 pKa = 11.56SLFSPMWIQPSTFEE292 pKa = 4.55LPVEE296 pKa = 4.5SGEE299 pKa = 4.65SLTIQYY305 pKa = 9.53NQDD308 pKa = 2.93GSSSVICGEE317 pKa = 3.97VEE319 pKa = 3.91AQIVRR324 pKa = 11.84SDD326 pKa = 2.88ITLNNGVMHH335 pKa = 7.53IIDD338 pKa = 4.1QILYY342 pKa = 6.48PTSTSSSSDD351 pKa = 3.13SAKK354 pKa = 10.64SNKK357 pKa = 7.87PTTTSSIPSTADD369 pKa = 2.95STSASIPDD377 pKa = 3.92GSDD380 pKa = 2.99SDD382 pKa = 4.24GQGGKK387 pKa = 9.87AAIPDD392 pKa = 3.84APQNSSTSSTVTLSVTLLGCLAVGWLVVYY421 pKa = 8.3VQQ423 pKa = 3.77
MM1 pKa = 7.05QLSIIPFLTILSILFPTSSAQLDD24 pKa = 3.8TDD26 pKa = 4.52IICPDD31 pKa = 3.4GSLTEE36 pKa = 4.4YY37 pKa = 8.58LTSMMDD43 pKa = 3.25VLFANGLTTFEE54 pKa = 3.89QLIVHH59 pKa = 6.61WSEE62 pKa = 3.22TDD64 pKa = 2.81MGYY67 pKa = 10.75EE68 pKa = 4.14FLHH71 pKa = 6.74DD72 pKa = 5.3LYY74 pKa = 11.73NSGQKK79 pKa = 8.99LTVLVPTNDD88 pKa = 3.19AFQEE92 pKa = 4.19SGIVSPFEE100 pKa = 4.18GLTEE104 pKa = 4.05DD105 pKa = 3.22WGAEE109 pKa = 3.88LGEE112 pKa = 4.01LHH114 pKa = 7.25LLQGEE119 pKa = 4.25WTYY122 pKa = 11.83DD123 pKa = 3.34QIPQSGHH130 pKa = 5.49AVAATSLLLANEE142 pKa = 4.47LNSTDD147 pKa = 3.58IQPNAYY153 pKa = 9.27QAMVLEE159 pKa = 4.82RR160 pKa = 11.84GNDD163 pKa = 3.42DD164 pKa = 3.61SVVVNGWWGHH174 pKa = 4.69ATSTSGPLDD183 pKa = 3.59LSGSGGLLDD192 pKa = 4.75NLFILPIDD200 pKa = 3.73QVLSFPPSLSTALQAPGLTNMSSALQVIGKK230 pKa = 9.76SSDD233 pKa = 3.43VEE235 pKa = 4.1RR236 pKa = 11.84LSEE239 pKa = 4.11GGFTIFVPLDD249 pKa = 3.72SVWNDD254 pKa = 3.21EE255 pKa = 5.0LKK257 pKa = 10.88DD258 pKa = 3.57IMLDD262 pKa = 3.49GSKK265 pKa = 10.54APEE268 pKa = 3.87MIGNHH273 pKa = 5.15FTTSYY278 pKa = 11.56SLFSPMWIQPSTFEE292 pKa = 4.55LPVEE296 pKa = 4.5SGEE299 pKa = 4.65SLTIQYY305 pKa = 9.53NQDD308 pKa = 2.93GSSSVICGEE317 pKa = 3.97VEE319 pKa = 3.91AQIVRR324 pKa = 11.84SDD326 pKa = 2.88ITLNNGVMHH335 pKa = 7.53IIDD338 pKa = 4.1QILYY342 pKa = 6.48PTSTSSSSDD351 pKa = 3.13SAKK354 pKa = 10.64SNKK357 pKa = 7.87PTTTSSIPSTADD369 pKa = 2.95STSASIPDD377 pKa = 3.92GSDD380 pKa = 2.99SDD382 pKa = 4.24GQGGKK387 pKa = 9.87AAIPDD392 pKa = 3.84APQNSSTSSTVTLSVTLLGCLAVGWLVVYY421 pKa = 8.3VQQ423 pKa = 3.77
Molecular weight: 45.18 kDa
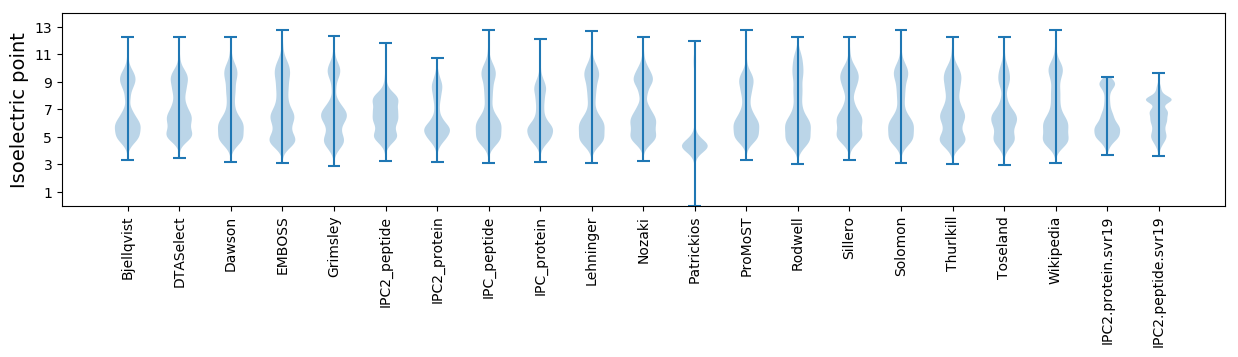
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B9IT04|A0A1B9IT04_9TREE Uncharacterized protein OS=Kwoniella mangroviensis CBS 10435 OX=1331196 GN=L486_03119 PE=3 SV=1
MM1 pKa = 7.16AQSEE5 pKa = 4.85VTQSLGLAIAQTEE18 pKa = 4.28NLFHH22 pKa = 6.68KK23 pKa = 10.59QADD26 pKa = 4.17LEE28 pKa = 4.3NSLNRR33 pKa = 11.84SLAISEE39 pKa = 4.48RR40 pKa = 11.84SLKK43 pKa = 10.24LANRR47 pKa = 11.84WVSSPSLSLFSGPSSMTLSSLGGVSTQLLIWSLQLIWQIAYY88 pKa = 10.77SMVSAFCFVFILFRR102 pKa = 11.84TGMRR106 pKa = 11.84KK107 pKa = 9.86SMLRR111 pKa = 11.84WSNRR115 pKa = 11.84FYY117 pKa = 10.69HH118 pKa = 6.35IPSDD122 pKa = 3.72EE123 pKa = 4.11EE124 pKa = 4.22SAVTSRR130 pKa = 11.84AKK132 pKa = 10.76DD133 pKa = 3.41FTNITISGITQSPSRR148 pKa = 11.84NRR150 pKa = 11.84QSCNPTANLAPSPEE164 pKa = 4.35VTRR167 pKa = 11.84TPDD170 pKa = 3.1PRR172 pKa = 11.84RR173 pKa = 11.84SNFRR177 pKa = 11.84RR178 pKa = 11.84SASAPLL184 pKa = 3.51
MM1 pKa = 7.16AQSEE5 pKa = 4.85VTQSLGLAIAQTEE18 pKa = 4.28NLFHH22 pKa = 6.68KK23 pKa = 10.59QADD26 pKa = 4.17LEE28 pKa = 4.3NSLNRR33 pKa = 11.84SLAISEE39 pKa = 4.48RR40 pKa = 11.84SLKK43 pKa = 10.24LANRR47 pKa = 11.84WVSSPSLSLFSGPSSMTLSSLGGVSTQLLIWSLQLIWQIAYY88 pKa = 10.77SMVSAFCFVFILFRR102 pKa = 11.84TGMRR106 pKa = 11.84KK107 pKa = 9.86SMLRR111 pKa = 11.84WSNRR115 pKa = 11.84FYY117 pKa = 10.69HH118 pKa = 6.35IPSDD122 pKa = 3.72EE123 pKa = 4.11EE124 pKa = 4.22SAVTSRR130 pKa = 11.84AKK132 pKa = 10.76DD133 pKa = 3.41FTNITISGITQSPSRR148 pKa = 11.84NRR150 pKa = 11.84QSCNPTANLAPSPEE164 pKa = 4.35VTRR167 pKa = 11.84TPDD170 pKa = 3.1PRR172 pKa = 11.84RR173 pKa = 11.84SNFRR177 pKa = 11.84RR178 pKa = 11.84SASAPLL184 pKa = 3.51
Molecular weight: 20.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4204977 |
50 |
4993 |
498.9 |
55.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.228 ± 0.029 | 0.972 ± 0.01 |
5.649 ± 0.018 | 6.465 ± 0.03 |
3.31 ± 0.015 | 7.093 ± 0.025 |
2.306 ± 0.011 | 5.263 ± 0.018 |
5.402 ± 0.024 | 8.765 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.052 ± 0.009 | 3.948 ± 0.016 |
6.554 ± 0.033 | 3.969 ± 0.021 |
5.682 ± 0.021 | 9.63 ± 0.039 |
6.104 ± 0.018 | 5.625 ± 0.019 |
1.344 ± 0.01 | 2.639 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
