
Torque teno midi virus 7
Taxonomy: Viruses; Anelloviridae; Gammatorquevirus
Average proteome isoelectric point is 7.77
Get precalculated fractions of proteins
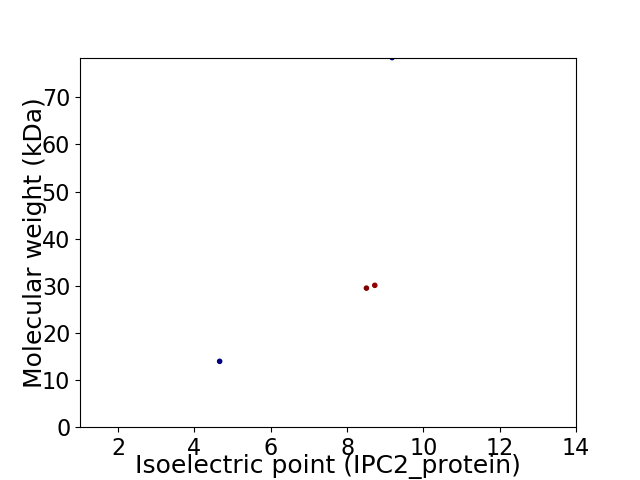
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
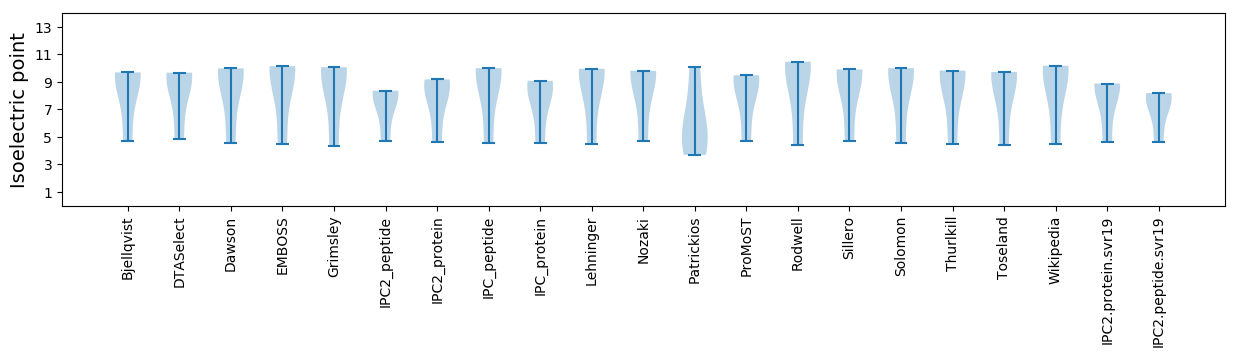
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A7VLW4|A7VLW4_9VIRU Capsid protein OS=Torque teno midi virus 7 OX=2065048 PE=3 SV=1
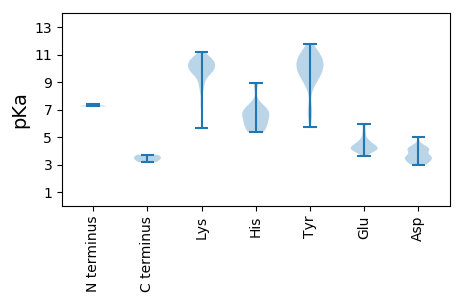
MM1 pKa = 7.23QNISADD7 pKa = 3.25NFFRR11 pKa = 11.84PTHH14 pKa = 6.07FNQVTKK20 pKa = 9.71QQIWMQQIQDD30 pKa = 3.49SHH32 pKa = 8.93DD33 pKa = 4.26NICNCNSCFAHH44 pKa = 6.84LLANIFPPGHH54 pKa = 7.21KK55 pKa = 10.53DD56 pKa = 3.0RR57 pKa = 11.84DD58 pKa = 3.49LSINQILARR67 pKa = 11.84DD68 pKa = 4.22LNQPCHH74 pKa = 6.46SGGTEE79 pKa = 3.77EE80 pKa = 4.75EE81 pKa = 4.39RR82 pKa = 11.84TGGDD86 pKa = 3.06TNTRR90 pKa = 11.84PEE92 pKa = 4.02NTGAADD98 pKa = 4.1GPAEE102 pKa = 4.09EE103 pKa = 4.55GKK105 pKa = 10.14HH106 pKa = 5.38YY107 pKa = 10.85IEE109 pKa = 5.94DD110 pKa = 3.89ADD112 pKa = 4.09LQEE115 pKa = 5.28LIAAGEE121 pKa = 4.21DD122 pKa = 3.18ATGRR126 pKa = 3.71
MM1 pKa = 7.23QNISADD7 pKa = 3.25NFFRR11 pKa = 11.84PTHH14 pKa = 6.07FNQVTKK20 pKa = 9.71QQIWMQQIQDD30 pKa = 3.49SHH32 pKa = 8.93DD33 pKa = 4.26NICNCNSCFAHH44 pKa = 6.84LLANIFPPGHH54 pKa = 7.21KK55 pKa = 10.53DD56 pKa = 3.0RR57 pKa = 11.84DD58 pKa = 3.49LSINQILARR67 pKa = 11.84DD68 pKa = 4.22LNQPCHH74 pKa = 6.46SGGTEE79 pKa = 3.77EE80 pKa = 4.75EE81 pKa = 4.39RR82 pKa = 11.84TGGDD86 pKa = 3.06TNTRR90 pKa = 11.84PEE92 pKa = 4.02NTGAADD98 pKa = 4.1GPAEE102 pKa = 4.09EE103 pKa = 4.55GKK105 pKa = 10.14HH106 pKa = 5.38YY107 pKa = 10.85IEE109 pKa = 5.94DD110 pKa = 3.89ADD112 pKa = 4.09LQEE115 pKa = 5.28LIAAGEE121 pKa = 4.21DD122 pKa = 3.18ATGRR126 pKa = 3.71
Molecular weight: 13.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A7VLW4|A7VLW4_9VIRU Capsid protein OS=Torque teno midi virus 7 OX=2065048 PE=3 SV=1
MM1 pKa = 7.42PFWWNRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 8.8NWWGYY15 pKa = 9.13KK16 pKa = 9.6YY17 pKa = 9.34KK18 pKa = 9.43TRR20 pKa = 11.84KK21 pKa = 7.44YY22 pKa = 10.27RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84WPRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84QTLYY36 pKa = 10.02RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84PTRR43 pKa = 11.84TNRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84HH52 pKa = 5.39RR53 pKa = 11.84KK54 pKa = 8.26VKK56 pKa = 10.24RR57 pKa = 11.84KK58 pKa = 9.99RR59 pKa = 11.84KK60 pKa = 7.35TLTVKK65 pKa = 9.7QWQPEE70 pKa = 4.42SIRR73 pKa = 11.84KK74 pKa = 9.01CKK76 pKa = 10.32IIGLEE81 pKa = 3.97PQLLGAEE88 pKa = 5.26GSQIDD93 pKa = 4.55CFTVVKK99 pKa = 9.82TDD101 pKa = 3.43YY102 pKa = 11.19VPAKK106 pKa = 9.64LPWGGGFSLQNYY118 pKa = 5.72TLKK121 pKa = 10.81YY122 pKa = 10.03LYY124 pKa = 10.32SEE126 pKa = 4.27YY127 pKa = 10.18TYY129 pKa = 11.13HH130 pKa = 6.63NNIWTSSNNQKK141 pKa = 10.36DD142 pKa = 3.57LCRR145 pKa = 11.84YY146 pKa = 6.72TGGYY150 pKa = 9.07IKK152 pKa = 10.7LFRR155 pKa = 11.84HH156 pKa = 5.43EE157 pKa = 4.26TQDD160 pKa = 4.32FIISYY165 pKa = 10.02DD166 pKa = 3.58LQPPHH171 pKa = 6.93EE172 pKa = 4.29LSKK175 pKa = 10.58YY176 pKa = 6.97TFPSCHH182 pKa = 5.8PHH184 pKa = 6.82QMLLQKK190 pKa = 9.92RR191 pKa = 11.84HH192 pKa = 6.49KK193 pKa = 10.17ILLSKK198 pKa = 10.57QSNPNGKK205 pKa = 9.49LYY207 pKa = 10.9KK208 pKa = 9.94KK209 pKa = 10.32IKK211 pKa = 9.99FKK213 pKa = 10.69PPKK216 pKa = 10.3QMINKK221 pKa = 8.7WLFTKK226 pKa = 10.69NFTPASLIVLKK237 pKa = 9.58TAVCNFSHH245 pKa = 7.22ANLSGKK251 pKa = 7.89NQNMLVSLISLNLSFYY267 pKa = 11.0KK268 pKa = 10.71NPAWAQALTASGYY281 pKa = 10.41KK282 pKa = 10.07PYY284 pKa = 11.28DD285 pKa = 4.28NIPSDD290 pKa = 3.83LPFVVKK296 pKa = 10.05QANKK300 pKa = 9.79EE301 pKa = 4.01VQKK304 pKa = 11.05YY305 pKa = 6.92MKK307 pKa = 9.88PQATTDD313 pKa = 3.5YY314 pKa = 8.87STSISYY320 pKa = 11.0DD321 pKa = 3.59DD322 pKa = 3.99GWFGPLFLSAIKK334 pKa = 10.03IGKK337 pKa = 8.85IEE339 pKa = 4.13QATATHH345 pKa = 6.29QIIQARR351 pKa = 11.84YY352 pKa = 8.51NPNIDD357 pKa = 3.65SGVGNQIYY365 pKa = 9.93CASTHH370 pKa = 6.24AYY372 pKa = 9.33SWAPPSKK379 pKa = 10.43DD380 pKa = 3.04KK381 pKa = 11.16QLLIEE386 pKa = 5.86GMPLWLGLFGYY397 pKa = 10.33YY398 pKa = 10.4SFVRR402 pKa = 11.84SVKK405 pKa = 10.7SEE407 pKa = 4.32DD408 pKa = 3.18FMKK411 pKa = 10.94AHH413 pKa = 6.97VICIKK418 pKa = 10.82SPALYY423 pKa = 9.46CYY425 pKa = 10.13PEE427 pKa = 5.59IGSCDD432 pKa = 3.27TYY434 pKa = 11.79VFIDD438 pKa = 3.69PEE440 pKa = 4.7FISGKK445 pKa = 9.91RR446 pKa = 11.84PYY448 pKa = 9.33EE449 pKa = 3.77QLITSQQKK457 pKa = 9.76RR458 pKa = 11.84LWYY461 pKa = 9.79PDD463 pKa = 3.32MSWQKK468 pKa = 9.78KK469 pKa = 5.66TMNTLVEE476 pKa = 4.44AGPYY480 pKa = 8.93VPKK483 pKa = 10.9LSDD486 pKa = 3.15EE487 pKa = 4.42TFSNWEE493 pKa = 4.0LNSRR497 pKa = 11.84YY498 pKa = 9.67CFYY501 pKa = 10.79FKK503 pKa = 10.24WGGPYY508 pKa = 9.46TDD510 pKa = 4.47EE511 pKa = 4.76PEE513 pKa = 4.05IANPEE518 pKa = 4.09MLPTYY523 pKa = 9.69DD524 pKa = 4.99VPDD527 pKa = 4.06TVQKK531 pKa = 9.62TIQIVNPEE539 pKa = 4.12KK540 pKa = 10.44QSPEE544 pKa = 4.29TIIHH548 pKa = 5.73PWDD551 pKa = 3.47YY552 pKa = 11.04RR553 pKa = 11.84RR554 pKa = 11.84GLIKK558 pKa = 10.45EE559 pKa = 4.03KK560 pKa = 10.44ALKK563 pKa = 10.34RR564 pKa = 11.84MCSNISTDD572 pKa = 3.55TEE574 pKa = 4.15FQPVAEE580 pKa = 4.43TTPKK584 pKa = 10.28KK585 pKa = 10.33KK586 pKa = 9.8PRR588 pKa = 11.84LRR590 pKa = 11.84ATLPNPEE597 pKa = 4.78EE598 pKa = 4.18EE599 pKa = 4.28TQEE602 pKa = 4.08VQKK605 pKa = 11.01CLLSLCEE612 pKa = 3.64EE613 pKa = 4.03STYY616 pKa = 10.5QEE618 pKa = 4.17EE619 pKa = 4.84EE620 pKa = 4.08EE621 pKa = 4.72TNLQQLIKK629 pKa = 8.57QQKK632 pKa = 8.2YY633 pKa = 6.77QQQKK637 pKa = 9.66LKK639 pKa = 10.73YY640 pKa = 9.52NMVKK644 pKa = 10.62LLIDD648 pKa = 4.09LKK650 pKa = 10.13EE651 pKa = 4.03KK652 pKa = 10.59QRR654 pKa = 11.84ILQQQTGLLDD664 pKa = 3.39
MM1 pKa = 7.42PFWWNRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 8.8NWWGYY15 pKa = 9.13KK16 pKa = 9.6YY17 pKa = 9.34KK18 pKa = 9.43TRR20 pKa = 11.84KK21 pKa = 7.44YY22 pKa = 10.27RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84WPRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84QTLYY36 pKa = 10.02RR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84PTRR43 pKa = 11.84TNRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84HH52 pKa = 5.39RR53 pKa = 11.84KK54 pKa = 8.26VKK56 pKa = 10.24RR57 pKa = 11.84KK58 pKa = 9.99RR59 pKa = 11.84KK60 pKa = 7.35TLTVKK65 pKa = 9.7QWQPEE70 pKa = 4.42SIRR73 pKa = 11.84KK74 pKa = 9.01CKK76 pKa = 10.32IIGLEE81 pKa = 3.97PQLLGAEE88 pKa = 5.26GSQIDD93 pKa = 4.55CFTVVKK99 pKa = 9.82TDD101 pKa = 3.43YY102 pKa = 11.19VPAKK106 pKa = 9.64LPWGGGFSLQNYY118 pKa = 5.72TLKK121 pKa = 10.81YY122 pKa = 10.03LYY124 pKa = 10.32SEE126 pKa = 4.27YY127 pKa = 10.18TYY129 pKa = 11.13HH130 pKa = 6.63NNIWTSSNNQKK141 pKa = 10.36DD142 pKa = 3.57LCRR145 pKa = 11.84YY146 pKa = 6.72TGGYY150 pKa = 9.07IKK152 pKa = 10.7LFRR155 pKa = 11.84HH156 pKa = 5.43EE157 pKa = 4.26TQDD160 pKa = 4.32FIISYY165 pKa = 10.02DD166 pKa = 3.58LQPPHH171 pKa = 6.93EE172 pKa = 4.29LSKK175 pKa = 10.58YY176 pKa = 6.97TFPSCHH182 pKa = 5.8PHH184 pKa = 6.82QMLLQKK190 pKa = 9.92RR191 pKa = 11.84HH192 pKa = 6.49KK193 pKa = 10.17ILLSKK198 pKa = 10.57QSNPNGKK205 pKa = 9.49LYY207 pKa = 10.9KK208 pKa = 9.94KK209 pKa = 10.32IKK211 pKa = 9.99FKK213 pKa = 10.69PPKK216 pKa = 10.3QMINKK221 pKa = 8.7WLFTKK226 pKa = 10.69NFTPASLIVLKK237 pKa = 9.58TAVCNFSHH245 pKa = 7.22ANLSGKK251 pKa = 7.89NQNMLVSLISLNLSFYY267 pKa = 11.0KK268 pKa = 10.71NPAWAQALTASGYY281 pKa = 10.41KK282 pKa = 10.07PYY284 pKa = 11.28DD285 pKa = 4.28NIPSDD290 pKa = 3.83LPFVVKK296 pKa = 10.05QANKK300 pKa = 9.79EE301 pKa = 4.01VQKK304 pKa = 11.05YY305 pKa = 6.92MKK307 pKa = 9.88PQATTDD313 pKa = 3.5YY314 pKa = 8.87STSISYY320 pKa = 11.0DD321 pKa = 3.59DD322 pKa = 3.99GWFGPLFLSAIKK334 pKa = 10.03IGKK337 pKa = 8.85IEE339 pKa = 4.13QATATHH345 pKa = 6.29QIIQARR351 pKa = 11.84YY352 pKa = 8.51NPNIDD357 pKa = 3.65SGVGNQIYY365 pKa = 9.93CASTHH370 pKa = 6.24AYY372 pKa = 9.33SWAPPSKK379 pKa = 10.43DD380 pKa = 3.04KK381 pKa = 11.16QLLIEE386 pKa = 5.86GMPLWLGLFGYY397 pKa = 10.33YY398 pKa = 10.4SFVRR402 pKa = 11.84SVKK405 pKa = 10.7SEE407 pKa = 4.32DD408 pKa = 3.18FMKK411 pKa = 10.94AHH413 pKa = 6.97VICIKK418 pKa = 10.82SPALYY423 pKa = 9.46CYY425 pKa = 10.13PEE427 pKa = 5.59IGSCDD432 pKa = 3.27TYY434 pKa = 11.79VFIDD438 pKa = 3.69PEE440 pKa = 4.7FISGKK445 pKa = 9.91RR446 pKa = 11.84PYY448 pKa = 9.33EE449 pKa = 3.77QLITSQQKK457 pKa = 9.76RR458 pKa = 11.84LWYY461 pKa = 9.79PDD463 pKa = 3.32MSWQKK468 pKa = 9.78KK469 pKa = 5.66TMNTLVEE476 pKa = 4.44AGPYY480 pKa = 8.93VPKK483 pKa = 10.9LSDD486 pKa = 3.15EE487 pKa = 4.42TFSNWEE493 pKa = 4.0LNSRR497 pKa = 11.84YY498 pKa = 9.67CFYY501 pKa = 10.79FKK503 pKa = 10.24WGGPYY508 pKa = 9.46TDD510 pKa = 4.47EE511 pKa = 4.76PEE513 pKa = 4.05IANPEE518 pKa = 4.09MLPTYY523 pKa = 9.69DD524 pKa = 4.99VPDD527 pKa = 4.06TVQKK531 pKa = 9.62TIQIVNPEE539 pKa = 4.12KK540 pKa = 10.44QSPEE544 pKa = 4.29TIIHH548 pKa = 5.73PWDD551 pKa = 3.47YY552 pKa = 11.04RR553 pKa = 11.84RR554 pKa = 11.84GLIKK558 pKa = 10.45EE559 pKa = 4.03KK560 pKa = 10.44ALKK563 pKa = 10.34RR564 pKa = 11.84MCSNISTDD572 pKa = 3.55TEE574 pKa = 4.15FQPVAEE580 pKa = 4.43TTPKK584 pKa = 10.28KK585 pKa = 10.33KK586 pKa = 9.8PRR588 pKa = 11.84LRR590 pKa = 11.84ATLPNPEE597 pKa = 4.78EE598 pKa = 4.18EE599 pKa = 4.28TQEE602 pKa = 4.08VQKK605 pKa = 11.01CLLSLCEE612 pKa = 3.64EE613 pKa = 4.03STYY616 pKa = 10.5QEE618 pKa = 4.17EE619 pKa = 4.84EE620 pKa = 4.08EE621 pKa = 4.72TNLQQLIKK629 pKa = 8.57QQKK632 pKa = 8.2YY633 pKa = 6.77QQQKK637 pKa = 9.66LKK639 pKa = 10.73YY640 pKa = 9.52NMVKK644 pKa = 10.62LLIDD648 pKa = 4.09LKK650 pKa = 10.13EE651 pKa = 4.03KK652 pKa = 10.59QRR654 pKa = 11.84ILQQQTGLLDD664 pKa = 3.39
Molecular weight: 78.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1321 |
126 |
664 |
330.3 |
38.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.602 ± 1.064 | 2.12 ± 0.232 |
4.542 ± 0.699 | 5.375 ± 0.396 |
3.709 ± 0.297 | 4.996 ± 0.591 |
2.801 ± 0.569 | 5.45 ± 0.402 |
8.478 ± 1.12 | 7.494 ± 0.7 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.665 ± 0.118 | 6.056 ± 0.714 |
6.813 ± 0.499 | 7.419 ± 0.765 |
5.753 ± 0.628 | 6.889 ± 1.162 |
6.813 ± 0.23 | 2.498 ± 0.644 |
1.817 ± 0.531 | 3.709 ± 1.331 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
