
Lake Sarah-associated circular molecule 10
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.56
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A126G8G1|A0A126G8G1_9VIRU Replication associated protein OS=Lake Sarah-associated circular molecule 10 OX=1685725 PE=4 SV=1
MM1 pKa = 6.78TTDD4 pKa = 5.19KK5 pKa = 11.24KK6 pKa = 10.43FTTQNNTKK14 pKa = 8.7YY15 pKa = 9.63WCFTWDD21 pKa = 3.38TNISQKK27 pKa = 10.69KK28 pKa = 8.72LPPVADD34 pKa = 5.41LINFLNNISEE44 pKa = 4.17TAIFQEE50 pKa = 4.19EE51 pKa = 4.91CGTIKK56 pKa = 10.84GKK58 pKa = 8.3VHH60 pKa = 5.26YY61 pKa = 10.15QGTLTLIGPRR71 pKa = 11.84QSKK74 pKa = 10.38ISVLRR79 pKa = 11.84LFEE82 pKa = 4.19RR83 pKa = 11.84RR84 pKa = 11.84FKK86 pKa = 10.96GVSGLTLSRR95 pKa = 11.84SYY97 pKa = 11.5SNQASEE103 pKa = 5.47DD104 pKa = 4.09YY105 pKa = 9.33VTKK108 pKa = 10.76DD109 pKa = 3.02EE110 pKa = 4.41GRR112 pKa = 11.84ISGPYY117 pKa = 9.45FCGKK121 pKa = 9.8KK122 pKa = 9.94EE123 pKa = 4.06KK124 pKa = 10.69FSMEE128 pKa = 3.86YY129 pKa = 9.37STTKK133 pKa = 9.16LTAWQQDD140 pKa = 3.55LFDD143 pKa = 5.22FLTKK147 pKa = 9.16ATKK150 pKa = 9.66IKK152 pKa = 10.14YY153 pKa = 9.05FRR155 pKa = 11.84DD156 pKa = 3.58RR157 pKa = 11.84IVIVVQDD164 pKa = 3.67EE165 pKa = 4.34KK166 pKa = 11.59GNSGKK171 pKa = 10.69SFFLKK176 pKa = 9.89WLAIGQKK183 pKa = 9.76VLNSKK188 pKa = 9.96KK189 pKa = 10.75LPVTSVDD196 pKa = 3.37RR197 pKa = 11.84LMSAVTKK204 pKa = 9.3VTADD208 pKa = 3.27SKK210 pKa = 11.2RR211 pKa = 11.84IDD213 pKa = 3.99LFTINLTRR221 pKa = 11.84SKK223 pKa = 11.42GEE225 pKa = 3.86DD226 pKa = 3.25QSYY229 pKa = 11.23KK230 pKa = 11.04DD231 pKa = 3.53LFAAIEE237 pKa = 4.33EE238 pKa = 4.45IKK240 pKa = 10.79DD241 pKa = 3.76GYY243 pKa = 10.39IVDD246 pKa = 4.13AMYY249 pKa = 11.23GNYY252 pKa = 9.52KK253 pKa = 9.85EE254 pKa = 5.82AIFDD258 pKa = 3.83PPIVVIFTNLIIDD271 pKa = 4.54GKK273 pKa = 10.38LSEE276 pKa = 4.67SLSPDD281 pKa = 2.45RR282 pKa = 11.84WLKK285 pKa = 9.58LTIDD289 pKa = 3.45NKK291 pKa = 11.11KK292 pKa = 9.9NIVYY296 pKa = 10.69NGIAWNAYY304 pKa = 7.17ITEE307 pKa = 4.39TPLKK311 pKa = 10.31DD312 pKa = 3.28IEE314 pKa = 4.35PEE316 pKa = 3.87EE317 pKa = 4.1FAKK320 pKa = 10.87NLSTKK325 pKa = 10.65YY326 pKa = 10.25LGEE329 pKa = 4.1KK330 pKa = 9.9
MM1 pKa = 6.78TTDD4 pKa = 5.19KK5 pKa = 11.24KK6 pKa = 10.43FTTQNNTKK14 pKa = 8.7YY15 pKa = 9.63WCFTWDD21 pKa = 3.38TNISQKK27 pKa = 10.69KK28 pKa = 8.72LPPVADD34 pKa = 5.41LINFLNNISEE44 pKa = 4.17TAIFQEE50 pKa = 4.19EE51 pKa = 4.91CGTIKK56 pKa = 10.84GKK58 pKa = 8.3VHH60 pKa = 5.26YY61 pKa = 10.15QGTLTLIGPRR71 pKa = 11.84QSKK74 pKa = 10.38ISVLRR79 pKa = 11.84LFEE82 pKa = 4.19RR83 pKa = 11.84RR84 pKa = 11.84FKK86 pKa = 10.96GVSGLTLSRR95 pKa = 11.84SYY97 pKa = 11.5SNQASEE103 pKa = 5.47DD104 pKa = 4.09YY105 pKa = 9.33VTKK108 pKa = 10.76DD109 pKa = 3.02EE110 pKa = 4.41GRR112 pKa = 11.84ISGPYY117 pKa = 9.45FCGKK121 pKa = 9.8KK122 pKa = 9.94EE123 pKa = 4.06KK124 pKa = 10.69FSMEE128 pKa = 3.86YY129 pKa = 9.37STTKK133 pKa = 9.16LTAWQQDD140 pKa = 3.55LFDD143 pKa = 5.22FLTKK147 pKa = 9.16ATKK150 pKa = 9.66IKK152 pKa = 10.14YY153 pKa = 9.05FRR155 pKa = 11.84DD156 pKa = 3.58RR157 pKa = 11.84IVIVVQDD164 pKa = 3.67EE165 pKa = 4.34KK166 pKa = 11.59GNSGKK171 pKa = 10.69SFFLKK176 pKa = 9.89WLAIGQKK183 pKa = 9.76VLNSKK188 pKa = 9.96KK189 pKa = 10.75LPVTSVDD196 pKa = 3.37RR197 pKa = 11.84LMSAVTKK204 pKa = 9.3VTADD208 pKa = 3.27SKK210 pKa = 11.2RR211 pKa = 11.84IDD213 pKa = 3.99LFTINLTRR221 pKa = 11.84SKK223 pKa = 11.42GEE225 pKa = 3.86DD226 pKa = 3.25QSYY229 pKa = 11.23KK230 pKa = 11.04DD231 pKa = 3.53LFAAIEE237 pKa = 4.33EE238 pKa = 4.45IKK240 pKa = 10.79DD241 pKa = 3.76GYY243 pKa = 10.39IVDD246 pKa = 4.13AMYY249 pKa = 11.23GNYY252 pKa = 9.52KK253 pKa = 9.85EE254 pKa = 5.82AIFDD258 pKa = 3.83PPIVVIFTNLIIDD271 pKa = 4.54GKK273 pKa = 10.38LSEE276 pKa = 4.67SLSPDD281 pKa = 2.45RR282 pKa = 11.84WLKK285 pKa = 9.58LTIDD289 pKa = 3.45NKK291 pKa = 11.11KK292 pKa = 9.9NIVYY296 pKa = 10.69NGIAWNAYY304 pKa = 7.17ITEE307 pKa = 4.39TPLKK311 pKa = 10.31DD312 pKa = 3.28IEE314 pKa = 4.35PEE316 pKa = 3.87EE317 pKa = 4.1FAKK320 pKa = 10.87NLSTKK325 pKa = 10.65YY326 pKa = 10.25LGEE329 pKa = 4.1KK330 pKa = 9.9
Molecular weight: 37.77 kDa
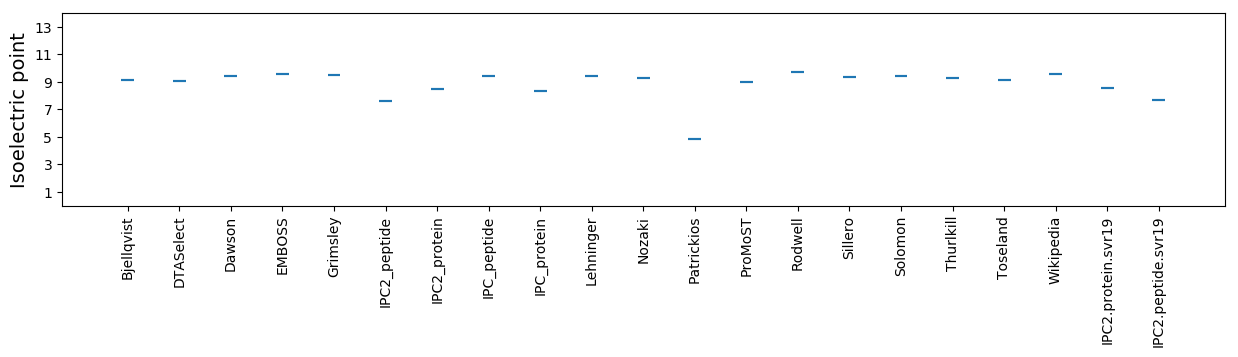
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A126G8G1|A0A126G8G1_9VIRU Replication associated protein OS=Lake Sarah-associated circular molecule 10 OX=1685725 PE=4 SV=1
MM1 pKa = 6.78TTDD4 pKa = 5.19KK5 pKa = 11.24KK6 pKa = 10.43FTTQNNTKK14 pKa = 8.7YY15 pKa = 9.63WCFTWDD21 pKa = 3.38TNISQKK27 pKa = 10.69KK28 pKa = 8.72LPPVADD34 pKa = 5.41LINFLNNISEE44 pKa = 4.17TAIFQEE50 pKa = 4.19EE51 pKa = 4.91CGTIKK56 pKa = 10.84GKK58 pKa = 8.3VHH60 pKa = 5.26YY61 pKa = 10.15QGTLTLIGPRR71 pKa = 11.84QSKK74 pKa = 10.38ISVLRR79 pKa = 11.84LFEE82 pKa = 4.19RR83 pKa = 11.84RR84 pKa = 11.84FKK86 pKa = 10.96GVSGLTLSRR95 pKa = 11.84SYY97 pKa = 11.5SNQASEE103 pKa = 5.47DD104 pKa = 4.09YY105 pKa = 9.33VTKK108 pKa = 10.76DD109 pKa = 3.02EE110 pKa = 4.41GRR112 pKa = 11.84ISGPYY117 pKa = 9.45FCGKK121 pKa = 9.8KK122 pKa = 9.94EE123 pKa = 4.06KK124 pKa = 10.69FSMEE128 pKa = 3.86YY129 pKa = 9.37STTKK133 pKa = 9.16LTAWQQDD140 pKa = 3.55LFDD143 pKa = 5.22FLTKK147 pKa = 9.16ATKK150 pKa = 9.66IKK152 pKa = 10.14YY153 pKa = 9.05FRR155 pKa = 11.84DD156 pKa = 3.58RR157 pKa = 11.84IVIVVQDD164 pKa = 3.67EE165 pKa = 4.34KK166 pKa = 11.59GNSGKK171 pKa = 10.69SFFLKK176 pKa = 9.89WLAIGQKK183 pKa = 9.76VLNSKK188 pKa = 9.96KK189 pKa = 10.75LPVTSVDD196 pKa = 3.37RR197 pKa = 11.84LMSAVTKK204 pKa = 9.3VTADD208 pKa = 3.27SKK210 pKa = 11.2RR211 pKa = 11.84IDD213 pKa = 3.99LFTINLTRR221 pKa = 11.84SKK223 pKa = 11.42GEE225 pKa = 3.86DD226 pKa = 3.25QSYY229 pKa = 11.23KK230 pKa = 11.04DD231 pKa = 3.53LFAAIEE237 pKa = 4.33EE238 pKa = 4.45IKK240 pKa = 10.79DD241 pKa = 3.76GYY243 pKa = 10.39IVDD246 pKa = 4.13AMYY249 pKa = 11.23GNYY252 pKa = 9.52KK253 pKa = 9.85EE254 pKa = 5.82AIFDD258 pKa = 3.83PPIVVIFTNLIIDD271 pKa = 4.54GKK273 pKa = 10.38LSEE276 pKa = 4.67SLSPDD281 pKa = 2.45RR282 pKa = 11.84WLKK285 pKa = 9.58LTIDD289 pKa = 3.45NKK291 pKa = 11.11KK292 pKa = 9.9NIVYY296 pKa = 10.69NGIAWNAYY304 pKa = 7.17ITEE307 pKa = 4.39TPLKK311 pKa = 10.31DD312 pKa = 3.28IEE314 pKa = 4.35PEE316 pKa = 3.87EE317 pKa = 4.1FAKK320 pKa = 10.87NLSTKK325 pKa = 10.65YY326 pKa = 10.25LGEE329 pKa = 4.1KK330 pKa = 9.9
MM1 pKa = 6.78TTDD4 pKa = 5.19KK5 pKa = 11.24KK6 pKa = 10.43FTTQNNTKK14 pKa = 8.7YY15 pKa = 9.63WCFTWDD21 pKa = 3.38TNISQKK27 pKa = 10.69KK28 pKa = 8.72LPPVADD34 pKa = 5.41LINFLNNISEE44 pKa = 4.17TAIFQEE50 pKa = 4.19EE51 pKa = 4.91CGTIKK56 pKa = 10.84GKK58 pKa = 8.3VHH60 pKa = 5.26YY61 pKa = 10.15QGTLTLIGPRR71 pKa = 11.84QSKK74 pKa = 10.38ISVLRR79 pKa = 11.84LFEE82 pKa = 4.19RR83 pKa = 11.84RR84 pKa = 11.84FKK86 pKa = 10.96GVSGLTLSRR95 pKa = 11.84SYY97 pKa = 11.5SNQASEE103 pKa = 5.47DD104 pKa = 4.09YY105 pKa = 9.33VTKK108 pKa = 10.76DD109 pKa = 3.02EE110 pKa = 4.41GRR112 pKa = 11.84ISGPYY117 pKa = 9.45FCGKK121 pKa = 9.8KK122 pKa = 9.94EE123 pKa = 4.06KK124 pKa = 10.69FSMEE128 pKa = 3.86YY129 pKa = 9.37STTKK133 pKa = 9.16LTAWQQDD140 pKa = 3.55LFDD143 pKa = 5.22FLTKK147 pKa = 9.16ATKK150 pKa = 9.66IKK152 pKa = 10.14YY153 pKa = 9.05FRR155 pKa = 11.84DD156 pKa = 3.58RR157 pKa = 11.84IVIVVQDD164 pKa = 3.67EE165 pKa = 4.34KK166 pKa = 11.59GNSGKK171 pKa = 10.69SFFLKK176 pKa = 9.89WLAIGQKK183 pKa = 9.76VLNSKK188 pKa = 9.96KK189 pKa = 10.75LPVTSVDD196 pKa = 3.37RR197 pKa = 11.84LMSAVTKK204 pKa = 9.3VTADD208 pKa = 3.27SKK210 pKa = 11.2RR211 pKa = 11.84IDD213 pKa = 3.99LFTINLTRR221 pKa = 11.84SKK223 pKa = 11.42GEE225 pKa = 3.86DD226 pKa = 3.25QSYY229 pKa = 11.23KK230 pKa = 11.04DD231 pKa = 3.53LFAAIEE237 pKa = 4.33EE238 pKa = 4.45IKK240 pKa = 10.79DD241 pKa = 3.76GYY243 pKa = 10.39IVDD246 pKa = 4.13AMYY249 pKa = 11.23GNYY252 pKa = 9.52KK253 pKa = 9.85EE254 pKa = 5.82AIFDD258 pKa = 3.83PPIVVIFTNLIIDD271 pKa = 4.54GKK273 pKa = 10.38LSEE276 pKa = 4.67SLSPDD281 pKa = 2.45RR282 pKa = 11.84WLKK285 pKa = 9.58LTIDD289 pKa = 3.45NKK291 pKa = 11.11KK292 pKa = 9.9NIVYY296 pKa = 10.69NGIAWNAYY304 pKa = 7.17ITEE307 pKa = 4.39TPLKK311 pKa = 10.31DD312 pKa = 3.28IEE314 pKa = 4.35PEE316 pKa = 3.87EE317 pKa = 4.1FAKK320 pKa = 10.87NLSTKK325 pKa = 10.65YY326 pKa = 10.25LGEE329 pKa = 4.1KK330 pKa = 9.9
Molecular weight: 37.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
330 |
330 |
330 |
330.0 |
37.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.545 ± 0.0 | 0.909 ± 0.0 |
6.364 ± 0.0 | 5.758 ± 0.0 |
5.455 ± 0.0 | 5.455 ± 0.0 |
0.303 ± 0.0 | 8.182 ± 0.0 |
11.212 ± 0.0 | 8.485 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.212 ± 0.0 | 5.152 ± 0.0 |
3.03 ± 0.0 | 3.333 ± 0.0 |
3.636 ± 0.0 | 7.273 ± 0.0 |
8.485 ± 0.0 | 5.152 ± 0.0 |
1.818 ± 0.0 | 4.242 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
