
Lichtheimia corymbifera JMRC:FSU:9682
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Fungi incertae sedis; Mucoromycota; Mucoromycotina; Mucoromycetes; Mucorales; Lichtheimiaceae; Lichtheimia; Lichtheimia corymbifera
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
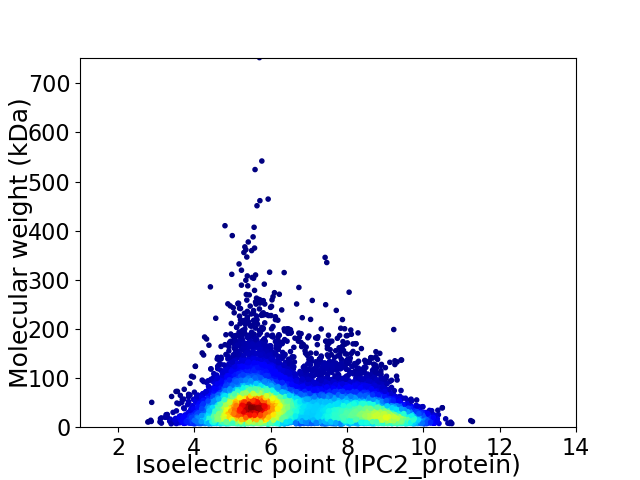
Virtual 2D-PAGE plot for 13257 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A068SC33|A0A068SC33_9FUNG Uncharacterized protein OS=Lichtheimia corymbifera JMRC:FSU:9682 OX=1263082 GN=LCOR_10216.1 PE=4 SV=1
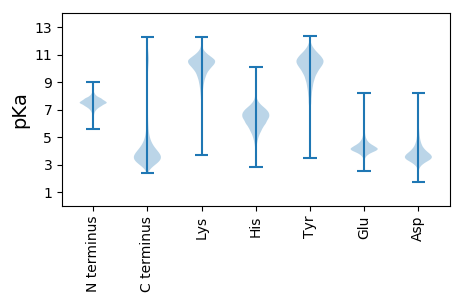
MM1 pKa = 7.86RR2 pKa = 11.84LLILLLILALAAMVFAQDD20 pKa = 3.92PGAGDD25 pKa = 3.54SGTGDD30 pKa = 3.29GVGGDD35 pKa = 3.71TGGDD39 pKa = 3.32TGSDD43 pKa = 3.28GSTGGDD49 pKa = 3.24GSTGAGDD56 pKa = 3.74GTTGGDD62 pKa = 3.24SGTGGGDD69 pKa = 3.09TSTGGDD75 pKa = 3.57GSTGGDD81 pKa = 3.44GSTGGDD87 pKa = 3.44GSTGGDD93 pKa = 3.41GSTGGGDD100 pKa = 3.32AGTGDD105 pKa = 4.11GSTGGDD111 pKa = 3.44TQQPSDD117 pKa = 4.85DD118 pKa = 4.46GSTTPNPTGDD128 pKa = 3.77GTTSPSPSDD137 pKa = 4.99DD138 pKa = 3.33GTTDD142 pKa = 3.31PQQPTDD148 pKa = 3.53QPSDD152 pKa = 3.17GGEE155 pKa = 3.98QQQPSPTDD163 pKa = 3.47PEE165 pKa = 4.32TAPDD169 pKa = 3.76ATPSGDD175 pKa = 3.75TVDD178 pKa = 4.54EE179 pKa = 4.55AACQALKK186 pKa = 10.56QIYY189 pKa = 10.16NSLNGNSWTVSNGWSGDD206 pKa = 3.76DD207 pKa = 3.75MTSCCNGWFGVSCIMGSVSKK227 pKa = 10.71IQLPYY232 pKa = 10.91NNLAGEE238 pKa = 4.32LPDD241 pKa = 4.17VFGSLQSLTNLDD253 pKa = 3.89LSHH256 pKa = 6.22NQISGSLPGSLTNMPNLRR274 pKa = 11.84FINVDD279 pKa = 3.41VNNLSGGLIAFGQTPMLQSIHH300 pKa = 6.03MKK302 pKa = 10.15NNSFTGSIPDD312 pKa = 2.89SWGNIGTLNDD322 pKa = 3.57IYY324 pKa = 11.12LSNNQLSGPFPMSILNLGQLTTLAIDD350 pKa = 3.35NNQFNGIVPPQIGNLGSLKK369 pKa = 10.48VLNIRR374 pKa = 11.84QNHH377 pKa = 5.85LLGNIPASIGNLTSLTNLLMSDD399 pKa = 3.18MMLNGTIPEE408 pKa = 4.17SLYY411 pKa = 11.21GLSNLQYY418 pKa = 11.25LDD420 pKa = 4.11LSHH423 pKa = 6.78NRR425 pKa = 11.84LTGEE429 pKa = 3.91ISDD432 pKa = 4.95KK433 pKa = 10.96LGQLVNVKK441 pKa = 10.52KK442 pKa = 11.0LMLGRR447 pKa = 11.84NGLTGPIPDD456 pKa = 4.18SLGALAQLEE465 pKa = 4.37YY466 pKa = 11.26VQLNYY471 pKa = 10.8NQLNGQFPSFMAPRR485 pKa = 11.84ALGYY489 pKa = 10.77CYY491 pKa = 7.47MTPNQFQLCPDD502 pKa = 3.64DD503 pKa = 4.88TIVHH507 pKa = 6.15NPEE510 pKa = 3.77SMAFQCATDD519 pKa = 4.03CFIGTTRR526 pKa = 11.84PGAGSSTRR534 pKa = 11.84ANLFVTIALVLLALFFYY551 pKa = 10.94CC552 pKa = 5.99
MM1 pKa = 7.86RR2 pKa = 11.84LLILLLILALAAMVFAQDD20 pKa = 3.92PGAGDD25 pKa = 3.54SGTGDD30 pKa = 3.29GVGGDD35 pKa = 3.71TGGDD39 pKa = 3.32TGSDD43 pKa = 3.28GSTGGDD49 pKa = 3.24GSTGAGDD56 pKa = 3.74GTTGGDD62 pKa = 3.24SGTGGGDD69 pKa = 3.09TSTGGDD75 pKa = 3.57GSTGGDD81 pKa = 3.44GSTGGDD87 pKa = 3.44GSTGGDD93 pKa = 3.41GSTGGGDD100 pKa = 3.32AGTGDD105 pKa = 4.11GSTGGDD111 pKa = 3.44TQQPSDD117 pKa = 4.85DD118 pKa = 4.46GSTTPNPTGDD128 pKa = 3.77GTTSPSPSDD137 pKa = 4.99DD138 pKa = 3.33GTTDD142 pKa = 3.31PQQPTDD148 pKa = 3.53QPSDD152 pKa = 3.17GGEE155 pKa = 3.98QQQPSPTDD163 pKa = 3.47PEE165 pKa = 4.32TAPDD169 pKa = 3.76ATPSGDD175 pKa = 3.75TVDD178 pKa = 4.54EE179 pKa = 4.55AACQALKK186 pKa = 10.56QIYY189 pKa = 10.16NSLNGNSWTVSNGWSGDD206 pKa = 3.76DD207 pKa = 3.75MTSCCNGWFGVSCIMGSVSKK227 pKa = 10.71IQLPYY232 pKa = 10.91NNLAGEE238 pKa = 4.32LPDD241 pKa = 4.17VFGSLQSLTNLDD253 pKa = 3.89LSHH256 pKa = 6.22NQISGSLPGSLTNMPNLRR274 pKa = 11.84FINVDD279 pKa = 3.41VNNLSGGLIAFGQTPMLQSIHH300 pKa = 6.03MKK302 pKa = 10.15NNSFTGSIPDD312 pKa = 2.89SWGNIGTLNDD322 pKa = 3.57IYY324 pKa = 11.12LSNNQLSGPFPMSILNLGQLTTLAIDD350 pKa = 3.35NNQFNGIVPPQIGNLGSLKK369 pKa = 10.48VLNIRR374 pKa = 11.84QNHH377 pKa = 5.85LLGNIPASIGNLTSLTNLLMSDD399 pKa = 3.18MMLNGTIPEE408 pKa = 4.17SLYY411 pKa = 11.21GLSNLQYY418 pKa = 11.25LDD420 pKa = 4.11LSHH423 pKa = 6.78NRR425 pKa = 11.84LTGEE429 pKa = 3.91ISDD432 pKa = 4.95KK433 pKa = 10.96LGQLVNVKK441 pKa = 10.52KK442 pKa = 11.0LMLGRR447 pKa = 11.84NGLTGPIPDD456 pKa = 4.18SLGALAQLEE465 pKa = 4.37YY466 pKa = 11.26VQLNYY471 pKa = 10.8NQLNGQFPSFMAPRR485 pKa = 11.84ALGYY489 pKa = 10.77CYY491 pKa = 7.47MTPNQFQLCPDD502 pKa = 3.64DD503 pKa = 4.88TIVHH507 pKa = 6.15NPEE510 pKa = 3.77SMAFQCATDD519 pKa = 4.03CFIGTTRR526 pKa = 11.84PGAGSSTRR534 pKa = 11.84ANLFVTIALVLLALFFYY551 pKa = 10.94CC552 pKa = 5.99
Molecular weight: 56.56 kDa
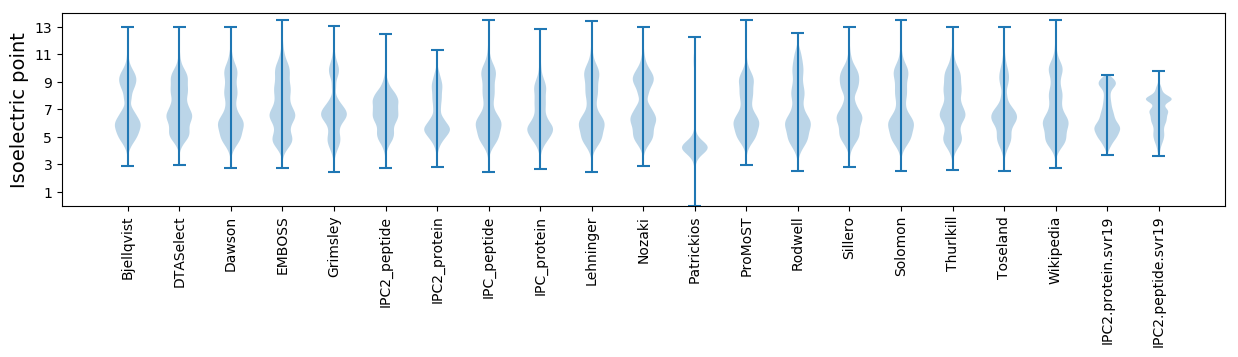
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A068RSL7|A0A068RSL7_9FUNG Uncharacterized protein OS=Lichtheimia corymbifera JMRC:FSU:9682 OX=1263082 GN=LCOR_04152.1 PE=4 SV=1
MM1 pKa = 7.26HH2 pKa = 7.14MKK4 pKa = 10.46GKK6 pKa = 8.7RR7 pKa = 11.84TRR9 pKa = 11.84TRR11 pKa = 11.84TRR13 pKa = 11.84TRR15 pKa = 11.84TRR17 pKa = 11.84THH19 pKa = 5.65ICAHH23 pKa = 5.6IRR25 pKa = 11.84AHH27 pKa = 5.83IRR29 pKa = 11.84THH31 pKa = 4.77IRR33 pKa = 11.84TRR35 pKa = 11.84TRR37 pKa = 11.84TRR39 pKa = 11.84TRR41 pKa = 11.84TRR43 pKa = 11.84THH45 pKa = 5.16STHH48 pKa = 5.75IRR50 pKa = 11.84THH52 pKa = 5.33IRR54 pKa = 11.84THH56 pKa = 5.44IRR58 pKa = 11.84THH60 pKa = 4.82IRR62 pKa = 11.84TRR64 pKa = 11.84TRR66 pKa = 11.84TRR68 pKa = 11.84TRR70 pKa = 11.84TRR72 pKa = 11.84THH74 pKa = 5.16STHH77 pKa = 5.75IRR79 pKa = 11.84THH81 pKa = 5.33IRR83 pKa = 11.84THH85 pKa = 5.44IRR87 pKa = 11.84THH89 pKa = 4.82IRR91 pKa = 11.84TRR93 pKa = 11.84TRR95 pKa = 11.84TRR97 pKa = 11.84TRR99 pKa = 11.84TRR101 pKa = 11.84TRR103 pKa = 11.84TRR105 pKa = 11.84SEE107 pKa = 3.71
MM1 pKa = 7.26HH2 pKa = 7.14MKK4 pKa = 10.46GKK6 pKa = 8.7RR7 pKa = 11.84TRR9 pKa = 11.84TRR11 pKa = 11.84TRR13 pKa = 11.84TRR15 pKa = 11.84TRR17 pKa = 11.84THH19 pKa = 5.65ICAHH23 pKa = 5.6IRR25 pKa = 11.84AHH27 pKa = 5.83IRR29 pKa = 11.84THH31 pKa = 4.77IRR33 pKa = 11.84TRR35 pKa = 11.84TRR37 pKa = 11.84TRR39 pKa = 11.84TRR41 pKa = 11.84TRR43 pKa = 11.84THH45 pKa = 5.16STHH48 pKa = 5.75IRR50 pKa = 11.84THH52 pKa = 5.33IRR54 pKa = 11.84THH56 pKa = 5.44IRR58 pKa = 11.84THH60 pKa = 4.82IRR62 pKa = 11.84TRR64 pKa = 11.84TRR66 pKa = 11.84TRR68 pKa = 11.84TRR70 pKa = 11.84TRR72 pKa = 11.84THH74 pKa = 5.16STHH77 pKa = 5.75IRR79 pKa = 11.84THH81 pKa = 5.33IRR83 pKa = 11.84THH85 pKa = 5.44IRR87 pKa = 11.84THH89 pKa = 4.82IRR91 pKa = 11.84TRR93 pKa = 11.84TRR95 pKa = 11.84TRR97 pKa = 11.84TRR99 pKa = 11.84TRR101 pKa = 11.84TRR103 pKa = 11.84TRR105 pKa = 11.84SEE107 pKa = 3.71
Molecular weight: 13.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5717124 |
66 |
6893 |
431.3 |
48.41 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.451 ± 0.019 | 1.446 ± 0.01 |
6.051 ± 0.016 | 5.815 ± 0.023 |
3.633 ± 0.014 | 5.404 ± 0.021 |
3.02 ± 0.014 | 5.333 ± 0.015 |
5.15 ± 0.021 | 8.791 ± 0.025 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.708 ± 0.009 | 4.241 ± 0.012 |
5.455 ± 0.025 | 4.967 ± 0.022 |
5.666 ± 0.018 | 8.255 ± 0.027 |
6.331 ± 0.018 | 5.878 ± 0.018 |
1.248 ± 0.007 | 3.154 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
