
Cybaeus spider associated circular virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses; Circularisvirus
Average proteome isoelectric point is 7.95
Get precalculated fractions of proteins
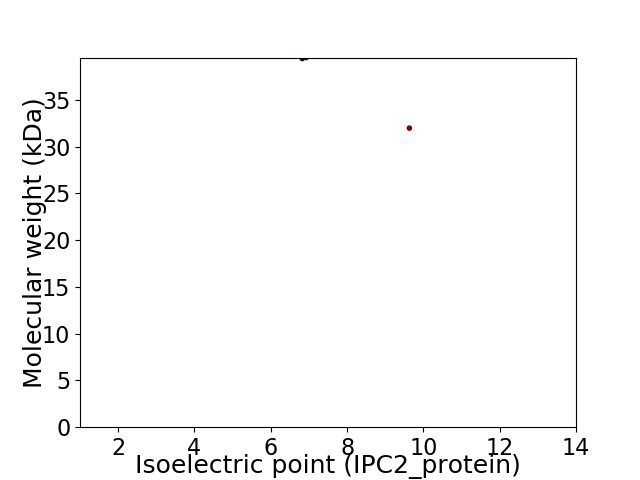
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
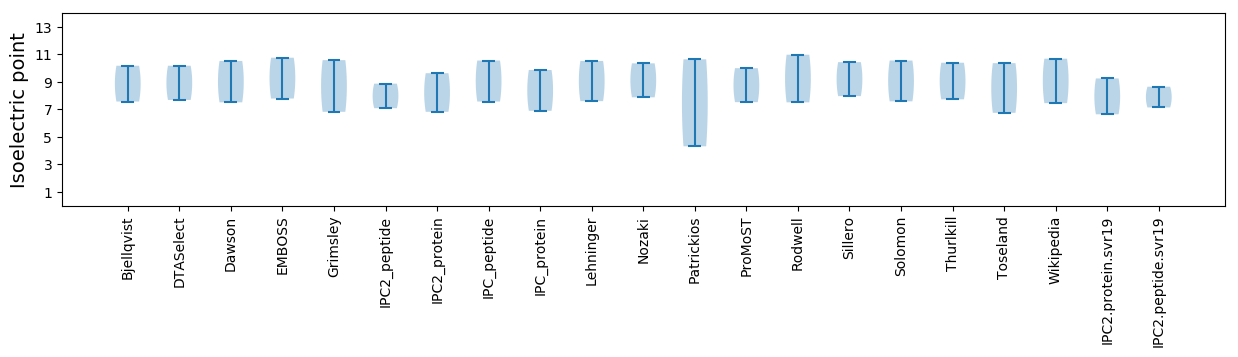
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPA9|A0A346BPA9_9VIRU Putative capsid protein OS=Cybaeus spider associated circular virus 1 OX=2293277 PE=4 SV=1
MM1 pKa = 7.73NSRR4 pKa = 11.84SKK6 pKa = 10.22NWCFTSYY13 pKa = 10.77NVEE16 pKa = 4.19SEE18 pKa = 4.38PFLQQRR24 pKa = 11.84NKK26 pKa = 9.87IQYY29 pKa = 8.53LIWGRR34 pKa = 11.84EE35 pKa = 3.86TCPDD39 pKa = 3.34TNRR42 pKa = 11.84PHH44 pKa = 6.61LQGFVAFKK52 pKa = 10.92VRR54 pKa = 11.84TKK56 pKa = 10.73FSTLKK61 pKa = 10.47NFEE64 pKa = 4.14NFHH67 pKa = 5.77FEE69 pKa = 4.0KK70 pKa = 10.43TKK72 pKa = 9.79GTIHH76 pKa = 6.6EE77 pKa = 4.16AVEE80 pKa = 4.17YY81 pKa = 10.16CKK83 pKa = 10.56KK84 pKa = 10.85DD85 pKa = 3.2GDD87 pKa = 3.86FTEE90 pKa = 4.85YY91 pKa = 11.05GVLPSATGNNSAFKK105 pKa = 10.43QVLAFAKK112 pKa = 9.97KK113 pKa = 10.41GEE115 pKa = 3.87ISAIEE120 pKa = 4.3DD121 pKa = 3.79EE122 pKa = 4.86HH123 pKa = 7.79PGIYY127 pKa = 9.42IRR129 pKa = 11.84YY130 pKa = 8.5KK131 pKa = 9.73RR132 pKa = 11.84TLEE135 pKa = 3.86SLAVYY140 pKa = 7.73NTSEE144 pKa = 4.57LEE146 pKa = 4.04NSCGVWIHH154 pKa = 6.14GPPRR158 pKa = 11.84CGKK161 pKa = 10.15DD162 pKa = 3.23YY163 pKa = 11.22AVRR166 pKa = 11.84QLGNVYY172 pKa = 10.55NKK174 pKa = 10.2GMNKK178 pKa = 8.32WWDD181 pKa = 3.53NYY183 pKa = 10.04KK184 pKa = 10.86GEE186 pKa = 4.83PIVLLSDD193 pKa = 3.67VQPDD197 pKa = 3.36QAKK200 pKa = 9.28WLGHH204 pKa = 5.63FLKK207 pKa = 10.28IWADD211 pKa = 3.78RR212 pKa = 11.84YY213 pKa = 11.07AFNAEE218 pKa = 4.28IKK220 pKa = 10.56CGTIYY225 pKa = 10.25IRR227 pKa = 11.84PKK229 pKa = 10.03QFYY232 pKa = 8.06VTSNFSIDD240 pKa = 3.71EE241 pKa = 4.04IFEE244 pKa = 4.23GKK246 pKa = 9.68IKK248 pKa = 10.25EE249 pKa = 4.34AIAARR254 pKa = 11.84FDD256 pKa = 3.73EE257 pKa = 5.44LDD259 pKa = 3.39FSTDD263 pKa = 2.87TNIIKK268 pKa = 10.34NRR270 pKa = 11.84LEE272 pKa = 4.34SIAPRR277 pKa = 11.84RR278 pKa = 11.84VLACLLDD285 pKa = 3.73RR286 pKa = 11.84QNIVAPAAVEE296 pKa = 4.43VTSDD300 pKa = 3.44PQPFQLHH307 pKa = 7.01DD308 pKa = 3.98GFSSEE313 pKa = 4.23MATAAATGVWSEE325 pKa = 4.92DD326 pKa = 3.29EE327 pKa = 5.4DD328 pKa = 4.13FVSPSFFTSQRR339 pKa = 11.84KK340 pKa = 8.17PKK342 pKa = 10.58NKK344 pKa = 10.13
MM1 pKa = 7.73NSRR4 pKa = 11.84SKK6 pKa = 10.22NWCFTSYY13 pKa = 10.77NVEE16 pKa = 4.19SEE18 pKa = 4.38PFLQQRR24 pKa = 11.84NKK26 pKa = 9.87IQYY29 pKa = 8.53LIWGRR34 pKa = 11.84EE35 pKa = 3.86TCPDD39 pKa = 3.34TNRR42 pKa = 11.84PHH44 pKa = 6.61LQGFVAFKK52 pKa = 10.92VRR54 pKa = 11.84TKK56 pKa = 10.73FSTLKK61 pKa = 10.47NFEE64 pKa = 4.14NFHH67 pKa = 5.77FEE69 pKa = 4.0KK70 pKa = 10.43TKK72 pKa = 9.79GTIHH76 pKa = 6.6EE77 pKa = 4.16AVEE80 pKa = 4.17YY81 pKa = 10.16CKK83 pKa = 10.56KK84 pKa = 10.85DD85 pKa = 3.2GDD87 pKa = 3.86FTEE90 pKa = 4.85YY91 pKa = 11.05GVLPSATGNNSAFKK105 pKa = 10.43QVLAFAKK112 pKa = 9.97KK113 pKa = 10.41GEE115 pKa = 3.87ISAIEE120 pKa = 4.3DD121 pKa = 3.79EE122 pKa = 4.86HH123 pKa = 7.79PGIYY127 pKa = 9.42IRR129 pKa = 11.84YY130 pKa = 8.5KK131 pKa = 9.73RR132 pKa = 11.84TLEE135 pKa = 3.86SLAVYY140 pKa = 7.73NTSEE144 pKa = 4.57LEE146 pKa = 4.04NSCGVWIHH154 pKa = 6.14GPPRR158 pKa = 11.84CGKK161 pKa = 10.15DD162 pKa = 3.23YY163 pKa = 11.22AVRR166 pKa = 11.84QLGNVYY172 pKa = 10.55NKK174 pKa = 10.2GMNKK178 pKa = 8.32WWDD181 pKa = 3.53NYY183 pKa = 10.04KK184 pKa = 10.86GEE186 pKa = 4.83PIVLLSDD193 pKa = 3.67VQPDD197 pKa = 3.36QAKK200 pKa = 9.28WLGHH204 pKa = 5.63FLKK207 pKa = 10.28IWADD211 pKa = 3.78RR212 pKa = 11.84YY213 pKa = 11.07AFNAEE218 pKa = 4.28IKK220 pKa = 10.56CGTIYY225 pKa = 10.25IRR227 pKa = 11.84PKK229 pKa = 10.03QFYY232 pKa = 8.06VTSNFSIDD240 pKa = 3.71EE241 pKa = 4.04IFEE244 pKa = 4.23GKK246 pKa = 9.68IKK248 pKa = 10.25EE249 pKa = 4.34AIAARR254 pKa = 11.84FDD256 pKa = 3.73EE257 pKa = 5.44LDD259 pKa = 3.39FSTDD263 pKa = 2.87TNIIKK268 pKa = 10.34NRR270 pKa = 11.84LEE272 pKa = 4.34SIAPRR277 pKa = 11.84RR278 pKa = 11.84VLACLLDD285 pKa = 3.73RR286 pKa = 11.84QNIVAPAAVEE296 pKa = 4.43VTSDD300 pKa = 3.44PQPFQLHH307 pKa = 7.01DD308 pKa = 3.98GFSSEE313 pKa = 4.23MATAAATGVWSEE325 pKa = 4.92DD326 pKa = 3.29EE327 pKa = 5.4DD328 pKa = 4.13FVSPSFFTSQRR339 pKa = 11.84KK340 pKa = 8.17PKK342 pKa = 10.58NKK344 pKa = 10.13
Molecular weight: 39.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPA9|A0A346BPA9_9VIRU Putative capsid protein OS=Cybaeus spider associated circular virus 1 OX=2293277 PE=4 SV=1
MM1 pKa = 7.1VFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84WPRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84VFGRR17 pKa = 11.84KK18 pKa = 7.48MRR20 pKa = 11.84TSYY23 pKa = 10.55RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84SLRR29 pKa = 11.84RR30 pKa = 11.84KK31 pKa = 9.86GNRR34 pKa = 11.84KK35 pKa = 9.06INKK38 pKa = 8.5CAQLNTVFFKK48 pKa = 10.81DD49 pKa = 3.49VQLFTMSLDD58 pKa = 3.81KK59 pKa = 11.16DD60 pKa = 4.16DD61 pKa = 4.29NTPGEE66 pKa = 4.12LNAAHH71 pKa = 7.09FLHH74 pKa = 6.97EE75 pKa = 4.28IKK77 pKa = 10.28PRR79 pKa = 11.84EE80 pKa = 3.6FGTRR84 pKa = 11.84YY85 pKa = 10.72KK86 pKa = 11.11NMCAKK91 pKa = 9.86HH92 pKa = 6.39RR93 pKa = 11.84YY94 pKa = 8.95VKK96 pKa = 10.01FLKK99 pKa = 8.96WRR101 pKa = 11.84YY102 pKa = 8.77FIKK105 pKa = 10.52LFQVTYY111 pKa = 9.18DD112 pKa = 3.5TVLLKK117 pKa = 10.28KK118 pKa = 10.51DD119 pKa = 3.54PKK121 pKa = 10.89DD122 pKa = 3.41PNNAFLATPGINSISNLPFRR142 pKa = 11.84INWDD146 pKa = 3.48LNHH149 pKa = 7.19IYY151 pKa = 10.06TKK153 pKa = 10.83GVAISDD159 pKa = 3.75QFDD162 pKa = 3.72DD163 pKa = 4.41PNFKK167 pKa = 9.34TVHH170 pKa = 6.26AGMKK174 pKa = 9.85KK175 pKa = 9.91AVQFTFNVPKK185 pKa = 10.44HH186 pKa = 5.06LQHH189 pKa = 6.5WVDD192 pKa = 3.85GPTIGAIDD200 pKa = 3.75VKK202 pKa = 10.87QSIGRR207 pKa = 11.84YY208 pKa = 9.19LEE210 pKa = 3.72QATNSKK216 pKa = 10.27NFAAPRR222 pKa = 11.84YY223 pKa = 8.7FVGTASDD230 pKa = 3.48LMKK233 pKa = 10.6NQNEE237 pKa = 4.39IIYY240 pKa = 9.69SDD242 pKa = 3.47RR243 pKa = 11.84KK244 pKa = 10.07EE245 pKa = 3.93PPLRR249 pKa = 11.84YY250 pKa = 9.04FLYY253 pKa = 8.55IQSYY257 pKa = 10.05AYY259 pKa = 9.09CTFKK263 pKa = 11.13GLTEE267 pKa = 4.24TDD269 pKa = 3.26TT270 pKa = 3.89
MM1 pKa = 7.1VFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84WPRR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84VFGRR17 pKa = 11.84KK18 pKa = 7.48MRR20 pKa = 11.84TSYY23 pKa = 10.55RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84SLRR29 pKa = 11.84RR30 pKa = 11.84KK31 pKa = 9.86GNRR34 pKa = 11.84KK35 pKa = 9.06INKK38 pKa = 8.5CAQLNTVFFKK48 pKa = 10.81DD49 pKa = 3.49VQLFTMSLDD58 pKa = 3.81KK59 pKa = 11.16DD60 pKa = 4.16DD61 pKa = 4.29NTPGEE66 pKa = 4.12LNAAHH71 pKa = 7.09FLHH74 pKa = 6.97EE75 pKa = 4.28IKK77 pKa = 10.28PRR79 pKa = 11.84EE80 pKa = 3.6FGTRR84 pKa = 11.84YY85 pKa = 10.72KK86 pKa = 11.11NMCAKK91 pKa = 9.86HH92 pKa = 6.39RR93 pKa = 11.84YY94 pKa = 8.95VKK96 pKa = 10.01FLKK99 pKa = 8.96WRR101 pKa = 11.84YY102 pKa = 8.77FIKK105 pKa = 10.52LFQVTYY111 pKa = 9.18DD112 pKa = 3.5TVLLKK117 pKa = 10.28KK118 pKa = 10.51DD119 pKa = 3.54PKK121 pKa = 10.89DD122 pKa = 3.41PNNAFLATPGINSISNLPFRR142 pKa = 11.84INWDD146 pKa = 3.48LNHH149 pKa = 7.19IYY151 pKa = 10.06TKK153 pKa = 10.83GVAISDD159 pKa = 3.75QFDD162 pKa = 3.72DD163 pKa = 4.41PNFKK167 pKa = 9.34TVHH170 pKa = 6.26AGMKK174 pKa = 9.85KK175 pKa = 9.91AVQFTFNVPKK185 pKa = 10.44HH186 pKa = 5.06LQHH189 pKa = 6.5WVDD192 pKa = 3.85GPTIGAIDD200 pKa = 3.75VKK202 pKa = 10.87QSIGRR207 pKa = 11.84YY208 pKa = 9.19LEE210 pKa = 3.72QATNSKK216 pKa = 10.27NFAAPRR222 pKa = 11.84YY223 pKa = 8.7FVGTASDD230 pKa = 3.48LMKK233 pKa = 10.6NQNEE237 pKa = 4.39IIYY240 pKa = 9.69SDD242 pKa = 3.47RR243 pKa = 11.84KK244 pKa = 10.07EE245 pKa = 3.93PPLRR249 pKa = 11.84YY250 pKa = 9.04FLYY253 pKa = 8.55IQSYY257 pKa = 10.05AYY259 pKa = 9.09CTFKK263 pKa = 11.13GLTEE267 pKa = 4.24TDD269 pKa = 3.26TT270 pKa = 3.89
Molecular weight: 31.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
614 |
270 |
344 |
307.0 |
35.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.515 ± 0.607 | 1.629 ± 0.328 |
5.537 ± 0.246 | 5.212 ± 1.659 |
7.003 ± 0.256 | 5.049 ± 0.383 |
2.28 ± 0.198 | 5.7 ± 0.326 |
8.469 ± 0.5 | 6.352 ± 0.434 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.466 ± 0.479 | 6.352 ± 0.199 |
4.723 ± 0.058 | 3.746 ± 0.027 |
6.84 ± 1.532 | 5.537 ± 0.927 |
6.026 ± 0.406 | 5.375 ± 0.12 |
1.954 ± 0.3 | 4.235 ± 0.368 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
