
Deer faeces associated circular DNA virus 1
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.93
Get precalculated fractions of proteins
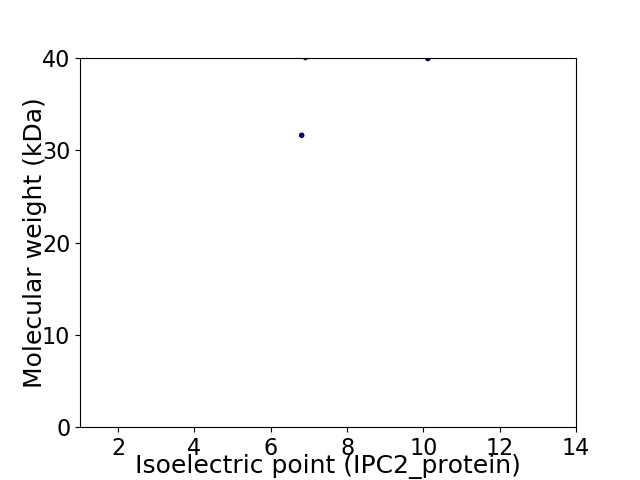
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
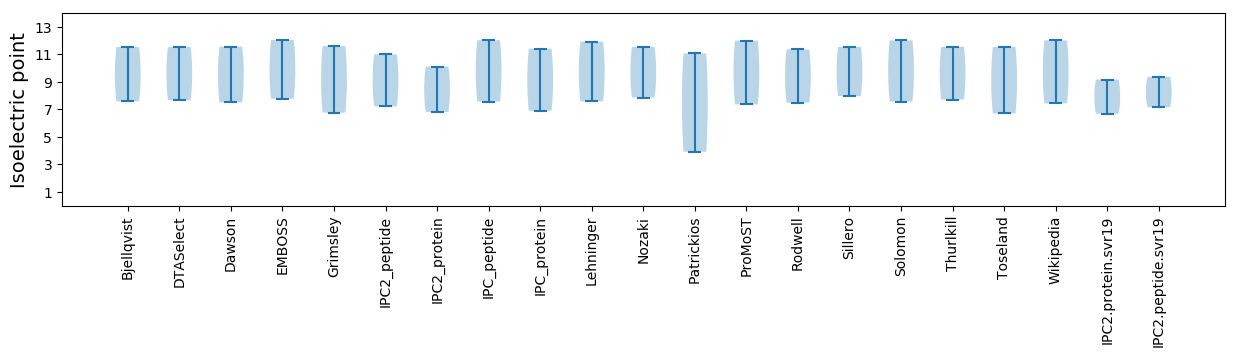
Summary statistics related to proteome-wise predictions
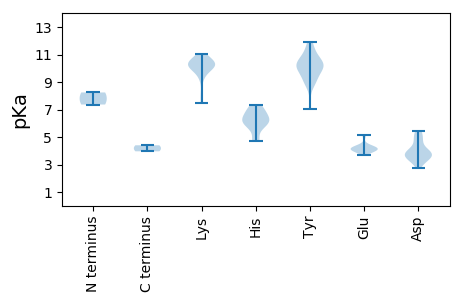
Protein with the lowest isoelectric point:
>tr|A0A161IBA2|A0A161IBA2_9VIRU Replication associated protein OS=Deer faeces associated circular DNA virus 1 OX=1843767 PE=4 SV=1
MM1 pKa = 8.24RR2 pKa = 11.84SRR4 pKa = 11.84TYY6 pKa = 10.43CYY8 pKa = 10.02TSFKK12 pKa = 10.31EE13 pKa = 4.03SEE15 pKa = 4.25PFVDD19 pKa = 3.07INQIVFQIHH28 pKa = 4.72QLEE31 pKa = 4.3ICEE34 pKa = 4.52TTGRR38 pKa = 11.84PHH40 pKa = 5.86WQGIVAFPNAKK51 pKa = 10.25APVGVKK57 pKa = 9.91QYY59 pKa = 11.37LCDD62 pKa = 3.49TSAHH66 pKa = 6.17IEE68 pKa = 4.01YY69 pKa = 9.01TKK71 pKa = 10.72NIPASIAYY79 pKa = 7.01CTKK82 pKa = 10.34EE83 pKa = 3.86EE84 pKa = 4.16TRR86 pKa = 11.84VDD88 pKa = 3.86GPFSYY93 pKa = 9.09GTVPDD98 pKa = 4.94LEE100 pKa = 4.99PDD102 pKa = 3.33NWWQQLSDD110 pKa = 3.55VQLWTEE116 pKa = 3.67HH117 pKa = 6.04SKK119 pKa = 10.48WMLRR123 pKa = 11.84NYY125 pKa = 10.55NGVAAFRR132 pKa = 11.84KK133 pKa = 9.84SIKK136 pKa = 9.97CVYY139 pKa = 10.06APRR142 pKa = 11.84ATPTVILLFGEE153 pKa = 4.41TGSGKK158 pKa = 10.11SRR160 pKa = 11.84AARR163 pKa = 11.84EE164 pKa = 3.87LAGDD168 pKa = 3.77EE169 pKa = 5.17LYY171 pKa = 10.01PKK173 pKa = 10.58PMGAWWDD180 pKa = 3.9GYY182 pKa = 9.91RR183 pKa = 11.84GHH185 pKa = 6.94RR186 pKa = 11.84VVLFDD191 pKa = 4.73DD192 pKa = 5.41FYY194 pKa = 11.9GNEE197 pKa = 4.06QYY199 pKa = 11.44SDD201 pKa = 3.68LLRR204 pKa = 11.84WLSEE208 pKa = 4.04LPISVPYY215 pKa = 10.38KK216 pKa = 10.84GGFTPLLADD225 pKa = 3.78RR226 pKa = 11.84FFFTSNAHH234 pKa = 5.82PRR236 pKa = 11.84DD237 pKa = 4.05WYY239 pKa = 10.56PNIPDD244 pKa = 4.71KK245 pKa = 11.05SALWRR250 pKa = 11.84RR251 pKa = 11.84VTKK254 pKa = 10.16IYY256 pKa = 10.94EE257 pKa = 4.16CFLDD261 pKa = 5.14RR262 pKa = 11.84FLICDD267 pKa = 3.9KK268 pKa = 10.91KK269 pKa = 10.59STLII273 pKa = 3.96
MM1 pKa = 8.24RR2 pKa = 11.84SRR4 pKa = 11.84TYY6 pKa = 10.43CYY8 pKa = 10.02TSFKK12 pKa = 10.31EE13 pKa = 4.03SEE15 pKa = 4.25PFVDD19 pKa = 3.07INQIVFQIHH28 pKa = 4.72QLEE31 pKa = 4.3ICEE34 pKa = 4.52TTGRR38 pKa = 11.84PHH40 pKa = 5.86WQGIVAFPNAKK51 pKa = 10.25APVGVKK57 pKa = 9.91QYY59 pKa = 11.37LCDD62 pKa = 3.49TSAHH66 pKa = 6.17IEE68 pKa = 4.01YY69 pKa = 9.01TKK71 pKa = 10.72NIPASIAYY79 pKa = 7.01CTKK82 pKa = 10.34EE83 pKa = 3.86EE84 pKa = 4.16TRR86 pKa = 11.84VDD88 pKa = 3.86GPFSYY93 pKa = 9.09GTVPDD98 pKa = 4.94LEE100 pKa = 4.99PDD102 pKa = 3.33NWWQQLSDD110 pKa = 3.55VQLWTEE116 pKa = 3.67HH117 pKa = 6.04SKK119 pKa = 10.48WMLRR123 pKa = 11.84NYY125 pKa = 10.55NGVAAFRR132 pKa = 11.84KK133 pKa = 9.84SIKK136 pKa = 9.97CVYY139 pKa = 10.06APRR142 pKa = 11.84ATPTVILLFGEE153 pKa = 4.41TGSGKK158 pKa = 10.11SRR160 pKa = 11.84AARR163 pKa = 11.84EE164 pKa = 3.87LAGDD168 pKa = 3.77EE169 pKa = 5.17LYY171 pKa = 10.01PKK173 pKa = 10.58PMGAWWDD180 pKa = 3.9GYY182 pKa = 9.91RR183 pKa = 11.84GHH185 pKa = 6.94RR186 pKa = 11.84VVLFDD191 pKa = 4.73DD192 pKa = 5.41FYY194 pKa = 11.9GNEE197 pKa = 4.06QYY199 pKa = 11.44SDD201 pKa = 3.68LLRR204 pKa = 11.84WLSEE208 pKa = 4.04LPISVPYY215 pKa = 10.38KK216 pKa = 10.84GGFTPLLADD225 pKa = 3.78RR226 pKa = 11.84FFFTSNAHH234 pKa = 5.82PRR236 pKa = 11.84DD237 pKa = 4.05WYY239 pKa = 10.56PNIPDD244 pKa = 4.71KK245 pKa = 11.05SALWRR250 pKa = 11.84RR251 pKa = 11.84VTKK254 pKa = 10.16IYY256 pKa = 10.94EE257 pKa = 4.16CFLDD261 pKa = 5.14RR262 pKa = 11.84FLICDD267 pKa = 3.9KK268 pKa = 10.91KK269 pKa = 10.59STLII273 pKa = 3.96
Molecular weight: 31.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A161IBA2|A0A161IBA2_9VIRU Replication associated protein OS=Deer faeces associated circular DNA virus 1 OX=1843767 PE=4 SV=1
MM1 pKa = 7.36SLGVIPRR8 pKa = 11.84VWPLIARR15 pKa = 11.84GIQRR19 pKa = 11.84LEE21 pKa = 4.17TYY23 pKa = 10.07PGGPRR28 pKa = 11.84GFAEE32 pKa = 3.9DD33 pKa = 3.48VVRR36 pKa = 11.84GVGYY40 pKa = 10.09GIQGYY45 pKa = 9.03KK46 pKa = 8.12VAKK49 pKa = 10.02NIAKK53 pKa = 10.09AYY55 pKa = 8.99GVHH58 pKa = 6.28KK59 pKa = 10.49VIEE62 pKa = 4.31AEE64 pKa = 4.01RR65 pKa = 11.84SVKK68 pKa = 10.16KK69 pKa = 10.24SRR71 pKa = 11.84KK72 pKa = 9.17NIQSIDD78 pKa = 3.43NPFMVKK84 pKa = 9.57RR85 pKa = 11.84RR86 pKa = 11.84FAGSIIRR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84PLRR101 pKa = 11.84RR102 pKa = 11.84PTHH105 pKa = 6.51RR106 pKa = 11.84FRR108 pKa = 11.84AGKK111 pKa = 10.04SRR113 pKa = 11.84FGHH116 pKa = 4.71GRR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84FGRR123 pKa = 11.84RR124 pKa = 11.84FGRR127 pKa = 11.84GGVKK131 pKa = 9.45RR132 pKa = 11.84VHH134 pKa = 6.14HH135 pKa = 7.31AIPTVTSRR143 pKa = 11.84EE144 pKa = 3.89FSMVTSFADD153 pKa = 3.92GARR156 pKa = 11.84YY157 pKa = 9.32LPLSIANSVTPSQFFQFFKK176 pKa = 10.35EE177 pKa = 4.15YY178 pKa = 10.22RR179 pKa = 11.84LEE181 pKa = 4.31RR182 pKa = 11.84CLVALRR188 pKa = 11.84RR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84PPCSVFEE198 pKa = 4.32SATGGNTVAAGRR210 pKa = 11.84EE211 pKa = 4.11EE212 pKa = 4.25TNDD215 pKa = 3.46IYY217 pKa = 10.47WVPWSTTDD225 pKa = 5.4LPVQHH230 pKa = 6.86PRR232 pKa = 11.84QIKK235 pKa = 7.45SARR238 pKa = 11.84MLTAMWSVCPIRR250 pKa = 11.84QRR252 pKa = 11.84CIEE255 pKa = 4.12MKK257 pKa = 10.54LINRR261 pKa = 11.84PLDD264 pKa = 3.85DD265 pKa = 4.37FLATIGGVPPNQIVTDD281 pKa = 4.01NSWAGTTQKK290 pKa = 10.43FPGYY294 pKa = 10.15AVQSWTRR301 pKa = 11.84MPWQQTDD308 pKa = 2.77ASLGDD313 pKa = 3.37RR314 pKa = 11.84HH315 pKa = 6.24LVHH318 pKa = 7.02VSQLLVLFLCLVLRR332 pKa = 11.84LFLVFRR338 pKa = 11.84ILIPFRR344 pKa = 11.84LTVFGG349 pKa = 4.4
MM1 pKa = 7.36SLGVIPRR8 pKa = 11.84VWPLIARR15 pKa = 11.84GIQRR19 pKa = 11.84LEE21 pKa = 4.17TYY23 pKa = 10.07PGGPRR28 pKa = 11.84GFAEE32 pKa = 3.9DD33 pKa = 3.48VVRR36 pKa = 11.84GVGYY40 pKa = 10.09GIQGYY45 pKa = 9.03KK46 pKa = 8.12VAKK49 pKa = 10.02NIAKK53 pKa = 10.09AYY55 pKa = 8.99GVHH58 pKa = 6.28KK59 pKa = 10.49VIEE62 pKa = 4.31AEE64 pKa = 4.01RR65 pKa = 11.84SVKK68 pKa = 10.16KK69 pKa = 10.24SRR71 pKa = 11.84KK72 pKa = 9.17NIQSIDD78 pKa = 3.43NPFMVKK84 pKa = 9.57RR85 pKa = 11.84RR86 pKa = 11.84FAGSIIRR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84PLRR101 pKa = 11.84RR102 pKa = 11.84PTHH105 pKa = 6.51RR106 pKa = 11.84FRR108 pKa = 11.84AGKK111 pKa = 10.04SRR113 pKa = 11.84FGHH116 pKa = 4.71GRR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84FGRR123 pKa = 11.84RR124 pKa = 11.84FGRR127 pKa = 11.84GGVKK131 pKa = 9.45RR132 pKa = 11.84VHH134 pKa = 6.14HH135 pKa = 7.31AIPTVTSRR143 pKa = 11.84EE144 pKa = 3.89FSMVTSFADD153 pKa = 3.92GARR156 pKa = 11.84YY157 pKa = 9.32LPLSIANSVTPSQFFQFFKK176 pKa = 10.35EE177 pKa = 4.15YY178 pKa = 10.22RR179 pKa = 11.84LEE181 pKa = 4.31RR182 pKa = 11.84CLVALRR188 pKa = 11.84RR189 pKa = 11.84RR190 pKa = 11.84RR191 pKa = 11.84PPCSVFEE198 pKa = 4.32SATGGNTVAAGRR210 pKa = 11.84EE211 pKa = 4.11EE212 pKa = 4.25TNDD215 pKa = 3.46IYY217 pKa = 10.47WVPWSTTDD225 pKa = 5.4LPVQHH230 pKa = 6.86PRR232 pKa = 11.84QIKK235 pKa = 7.45SARR238 pKa = 11.84MLTAMWSVCPIRR250 pKa = 11.84QRR252 pKa = 11.84CIEE255 pKa = 4.12MKK257 pKa = 10.54LINRR261 pKa = 11.84PLDD264 pKa = 3.85DD265 pKa = 4.37FLATIGGVPPNQIVTDD281 pKa = 4.01NSWAGTTQKK290 pKa = 10.43FPGYY294 pKa = 10.15AVQSWTRR301 pKa = 11.84MPWQQTDD308 pKa = 2.77ASLGDD313 pKa = 3.37RR314 pKa = 11.84HH315 pKa = 6.24LVHH318 pKa = 7.02VSQLLVLFLCLVLRR332 pKa = 11.84LFLVFRR338 pKa = 11.84ILIPFRR344 pKa = 11.84LTVFGG349 pKa = 4.4
Molecular weight: 39.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
622 |
273 |
349 |
311.0 |
35.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.27 ± 0.025 | 1.929 ± 0.372 |
4.18 ± 0.986 | 4.18 ± 0.771 |
5.788 ± 0.172 | 7.074 ± 0.712 |
2.251 ± 0.031 | 5.788 ± 0.172 |
4.662 ± 0.488 | 7.556 ± 0.08 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.608 ± 0.299 | 2.894 ± 0.236 |
6.431 ± 0.095 | 3.698 ± 0.235 |
10.129 ± 2.289 | 6.109 ± 0.069 |
5.788 ± 0.258 | 7.235 ± 1.021 |
2.733 ± 0.546 | 3.698 ± 1.054 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
