
Dishui Lake virophage 4
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Maveriviricetes; Priklausovirales; Lavidaviridae; unclassified Lavidaviridae
Average proteome isoelectric point is 7.11
Get precalculated fractions of proteins
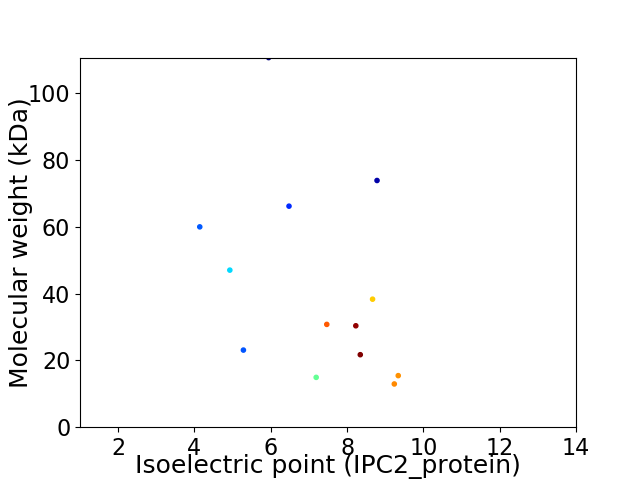
Virtual 2D-PAGE plot for 13 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6G6XNE5|A0A6G6XNE5_9VIRU RING-type domain-containing protein OS=Dishui Lake virophage 4 OX=2704065 PE=4 SV=1
MM1 pKa = 7.75SFGTFWGTDD10 pKa = 3.42PTDD13 pKa = 2.87VATYY17 pKa = 10.29QKK19 pKa = 10.78ILGKK23 pKa = 10.0YY24 pKa = 9.65LEE26 pKa = 4.93GGAAAVPPLNSVLGVGNSAANQSATDD52 pKa = 4.42FNTLGCIEE60 pKa = 4.42IEE62 pKa = 4.17TGKK65 pKa = 10.69VYY67 pKa = 10.6QGNNLDD73 pKa = 4.03LEE75 pKa = 4.51IGEE78 pKa = 4.64VGDD81 pKa = 3.58TLKK84 pKa = 10.84ILGAITKK91 pKa = 10.39GSILVGNGVEE101 pKa = 4.47TKK103 pKa = 9.82EE104 pKa = 4.51LPVGANGLVLKK115 pKa = 10.26ANSGAAYY122 pKa = 9.32GVEE125 pKa = 3.98WGTDD129 pKa = 3.23ASGGTVMAVNAGTNISVSGTIAQPIVNFATPTTSNIVLGVGTEE172 pKa = 3.98IEE174 pKa = 4.31ATNGATTMTLDD185 pKa = 3.33ATGLNDD191 pKa = 3.69TYY193 pKa = 11.67SSGGVVNQEE202 pKa = 4.28DD203 pKa = 4.02IEE205 pKa = 4.63VNATSVVQLLSATDD219 pKa = 3.37GGSYY223 pKa = 10.98LNTDD227 pKa = 3.5TTNVSNTGIVEE238 pKa = 4.11NLSATNLSTNNIGNVSLTCFSNSAGIACGCSAPTTPPYY276 pKa = 10.56PEE278 pKa = 4.09VSATAGLSATTTNAQLTISQSAPFAISYY306 pKa = 7.55STTLDD311 pKa = 3.18INGITQNNTGGVAGFTINTNTQPLALTTGDD341 pKa = 4.78HH342 pKa = 5.95ITFSADD348 pKa = 4.38NIDD351 pKa = 3.92LDD353 pKa = 3.74ATGRR357 pKa = 11.84LVLPSLASGDD367 pKa = 3.84YY368 pKa = 10.74LDD370 pKa = 5.15YY371 pKa = 11.34NAGSLKK377 pKa = 10.15IVNDD381 pKa = 3.89SVGGSANPLLVLQNNNNTAGATTFEE406 pKa = 4.78TYY408 pKa = 10.85KK409 pKa = 10.65NDD411 pKa = 3.34QPTSTGGDD419 pKa = 3.72NIASWSATCNTNVGKK434 pKa = 8.64TEE436 pKa = 3.72ISRR439 pKa = 11.84VNHH442 pKa = 5.78IAYY445 pKa = 7.64GTGASNNDD453 pKa = 3.12GGISLACKK461 pKa = 9.9VNSSITNFLICNGGANNAIGSGEE484 pKa = 4.09VQVFKK489 pKa = 10.54PITSTSPNNLNLICPTAGGTAKK511 pKa = 9.97IQSVSNIEE519 pKa = 4.38LEE521 pKa = 4.19ATGDD525 pKa = 3.61NLTTISGANTTISTTGVGSQIEE547 pKa = 4.52FKK549 pKa = 11.0PEE551 pKa = 3.31TTAGKK556 pKa = 9.98IVFTGASLQSNTSSGNSGEE575 pKa = 4.2HH576 pKa = 5.67LVIYY580 pKa = 10.46LNGVKK585 pKa = 9.75YY586 pKa = 10.24HH587 pKa = 7.04IALQTPP593 pKa = 4.14
MM1 pKa = 7.75SFGTFWGTDD10 pKa = 3.42PTDD13 pKa = 2.87VATYY17 pKa = 10.29QKK19 pKa = 10.78ILGKK23 pKa = 10.0YY24 pKa = 9.65LEE26 pKa = 4.93GGAAAVPPLNSVLGVGNSAANQSATDD52 pKa = 4.42FNTLGCIEE60 pKa = 4.42IEE62 pKa = 4.17TGKK65 pKa = 10.69VYY67 pKa = 10.6QGNNLDD73 pKa = 4.03LEE75 pKa = 4.51IGEE78 pKa = 4.64VGDD81 pKa = 3.58TLKK84 pKa = 10.84ILGAITKK91 pKa = 10.39GSILVGNGVEE101 pKa = 4.47TKK103 pKa = 9.82EE104 pKa = 4.51LPVGANGLVLKK115 pKa = 10.26ANSGAAYY122 pKa = 9.32GVEE125 pKa = 3.98WGTDD129 pKa = 3.23ASGGTVMAVNAGTNISVSGTIAQPIVNFATPTTSNIVLGVGTEE172 pKa = 3.98IEE174 pKa = 4.31ATNGATTMTLDD185 pKa = 3.33ATGLNDD191 pKa = 3.69TYY193 pKa = 11.67SSGGVVNQEE202 pKa = 4.28DD203 pKa = 4.02IEE205 pKa = 4.63VNATSVVQLLSATDD219 pKa = 3.37GGSYY223 pKa = 10.98LNTDD227 pKa = 3.5TTNVSNTGIVEE238 pKa = 4.11NLSATNLSTNNIGNVSLTCFSNSAGIACGCSAPTTPPYY276 pKa = 10.56PEE278 pKa = 4.09VSATAGLSATTTNAQLTISQSAPFAISYY306 pKa = 7.55STTLDD311 pKa = 3.18INGITQNNTGGVAGFTINTNTQPLALTTGDD341 pKa = 4.78HH342 pKa = 5.95ITFSADD348 pKa = 4.38NIDD351 pKa = 3.92LDD353 pKa = 3.74ATGRR357 pKa = 11.84LVLPSLASGDD367 pKa = 3.84YY368 pKa = 10.74LDD370 pKa = 5.15YY371 pKa = 11.34NAGSLKK377 pKa = 10.15IVNDD381 pKa = 3.89SVGGSANPLLVLQNNNNTAGATTFEE406 pKa = 4.78TYY408 pKa = 10.85KK409 pKa = 10.65NDD411 pKa = 3.34QPTSTGGDD419 pKa = 3.72NIASWSATCNTNVGKK434 pKa = 8.64TEE436 pKa = 3.72ISRR439 pKa = 11.84VNHH442 pKa = 5.78IAYY445 pKa = 7.64GTGASNNDD453 pKa = 3.12GGISLACKK461 pKa = 9.9VNSSITNFLICNGGANNAIGSGEE484 pKa = 4.09VQVFKK489 pKa = 10.54PITSTSPNNLNLICPTAGGTAKK511 pKa = 9.97IQSVSNIEE519 pKa = 4.38LEE521 pKa = 4.19ATGDD525 pKa = 3.61NLTTISGANTTISTTGVGSQIEE547 pKa = 4.52FKK549 pKa = 11.0PEE551 pKa = 3.31TTAGKK556 pKa = 9.98IVFTGASLQSNTSSGNSGEE575 pKa = 4.2HH576 pKa = 5.67LVIYY580 pKa = 10.46LNGVKK585 pKa = 9.75YY586 pKa = 10.24HH587 pKa = 7.04IALQTPP593 pKa = 4.14
Molecular weight: 60.02 kDa
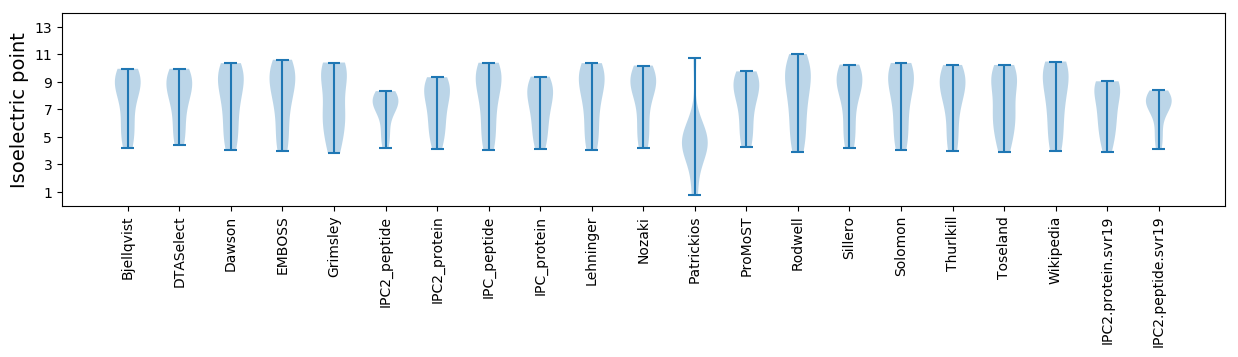
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6G6XMQ5|A0A6G6XMQ5_9VIRU Uncharacterized protein OS=Dishui Lake virophage 4 OX=2704065 PE=4 SV=1
MM1 pKa = 7.41EE2 pKa = 4.68RR3 pKa = 11.84RR4 pKa = 11.84FIFRR8 pKa = 11.84NYY10 pKa = 8.17KK11 pKa = 6.84TAKK14 pKa = 10.0KK15 pKa = 9.37YY16 pKa = 11.2GEE18 pKa = 4.52TIVPIPKK25 pKa = 9.47EE26 pKa = 3.76LHH28 pKa = 7.16DD29 pKa = 3.67ILAVYY34 pKa = 9.42FEE36 pKa = 4.68KK37 pKa = 10.72KK38 pKa = 10.39GILRR42 pKa = 11.84RR43 pKa = 11.84LQAPVKK49 pKa = 8.73KK50 pKa = 8.29TKK52 pKa = 10.36KK53 pKa = 8.32EE54 pKa = 3.52ASIFIEE60 pKa = 4.44PFLTLWNDD68 pKa = 3.03KK69 pKa = 10.34PFLINTITRR78 pKa = 11.84ILNRR82 pKa = 11.84VFGKK86 pKa = 10.66KK87 pKa = 9.32IGSSMLRR94 pKa = 11.84HH95 pKa = 6.69IYY97 pKa = 6.2TTKK100 pKa = 10.92KK101 pKa = 10.15FGKK104 pKa = 9.61QLAEE108 pKa = 3.92QKK110 pKa = 8.5EE111 pKa = 4.28TAEE114 pKa = 4.06QMGHH118 pKa = 5.79SVAEE122 pKa = 4.14MNQTYY127 pKa = 10.18IKK129 pKa = 10.52EE130 pKa = 4.17DD131 pKa = 3.15
MM1 pKa = 7.41EE2 pKa = 4.68RR3 pKa = 11.84RR4 pKa = 11.84FIFRR8 pKa = 11.84NYY10 pKa = 8.17KK11 pKa = 6.84TAKK14 pKa = 10.0KK15 pKa = 9.37YY16 pKa = 11.2GEE18 pKa = 4.52TIVPIPKK25 pKa = 9.47EE26 pKa = 3.76LHH28 pKa = 7.16DD29 pKa = 3.67ILAVYY34 pKa = 9.42FEE36 pKa = 4.68KK37 pKa = 10.72KK38 pKa = 10.39GILRR42 pKa = 11.84RR43 pKa = 11.84LQAPVKK49 pKa = 8.73KK50 pKa = 8.29TKK52 pKa = 10.36KK53 pKa = 8.32EE54 pKa = 3.52ASIFIEE60 pKa = 4.44PFLTLWNDD68 pKa = 3.03KK69 pKa = 10.34PFLINTITRR78 pKa = 11.84ILNRR82 pKa = 11.84VFGKK86 pKa = 10.66KK87 pKa = 9.32IGSSMLRR94 pKa = 11.84HH95 pKa = 6.69IYY97 pKa = 6.2TTKK100 pKa = 10.92KK101 pKa = 10.15FGKK104 pKa = 9.61QLAEE108 pKa = 3.92QKK110 pKa = 8.5EE111 pKa = 4.28TAEE114 pKa = 4.06QMGHH118 pKa = 5.79SVAEE122 pKa = 4.14MNQTYY127 pKa = 10.18IKK129 pKa = 10.52EE130 pKa = 4.17DD131 pKa = 3.15
Molecular weight: 15.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4887 |
114 |
963 |
375.9 |
41.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.469 ± 0.48 | 1.166 ± 0.188 |
5.463 ± 0.342 | 6.384 ± 0.814 |
3.458 ± 0.298 | 6.834 ± 0.735 |
1.228 ± 0.135 | 6.528 ± 0.547 |
7.285 ± 1.034 | 8.615 ± 0.296 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.415 ± 0.304 | 6.384 ± 0.568 |
4.931 ± 0.572 | 3.397 ± 0.22 |
4.87 ± 0.72 | 6.65 ± 0.924 |
6.077 ± 0.955 | 5.77 ± 0.241 |
1.187 ± 0.201 | 3.888 ± 0.33 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
