
Marinobacter sp. Z-F4-2
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Marinobacter; unclassified Marinobacter
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
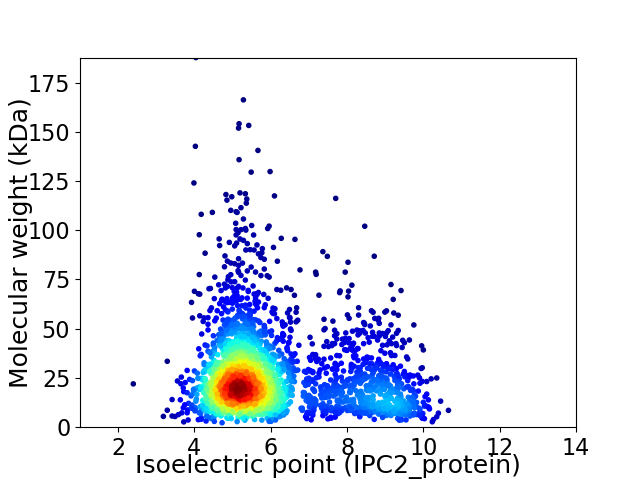
Virtual 2D-PAGE plot for 2568 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T4DBT8|A0A2T4DBT8_9ALTE Fatty acid-binding protein DegV (Fragment) OS=Marinobacter sp. Z-F4-2 OX=2137199 GN=C9993_12540 PE=4 SV=1
MM1 pKa = 7.74SDD3 pKa = 3.39FRR5 pKa = 11.84EE6 pKa = 4.16EE7 pKa = 5.22LEE9 pKa = 4.34GCQGNKK15 pKa = 10.18SLILGNGFGISYY27 pKa = 10.45DD28 pKa = 3.35VAARR32 pKa = 11.84TDD34 pKa = 3.76SFCWDD39 pKa = 3.79SLADD43 pKa = 4.67LCDD46 pKa = 4.09FDD48 pKa = 5.34NDD50 pKa = 3.71NPLLEE55 pKa = 5.0LLEE58 pKa = 4.24EE59 pKa = 4.62CNFDD63 pKa = 4.91FEE65 pKa = 5.41IVHH68 pKa = 5.8QKK70 pKa = 9.83INNAISVII78 pKa = 3.82
MM1 pKa = 7.74SDD3 pKa = 3.39FRR5 pKa = 11.84EE6 pKa = 4.16EE7 pKa = 5.22LEE9 pKa = 4.34GCQGNKK15 pKa = 10.18SLILGNGFGISYY27 pKa = 10.45DD28 pKa = 3.35VAARR32 pKa = 11.84TDD34 pKa = 3.76SFCWDD39 pKa = 3.79SLADD43 pKa = 4.67LCDD46 pKa = 4.09FDD48 pKa = 5.34NDD50 pKa = 3.71NPLLEE55 pKa = 5.0LLEE58 pKa = 4.24EE59 pKa = 4.62CNFDD63 pKa = 4.91FEE65 pKa = 5.41IVHH68 pKa = 5.8QKK70 pKa = 9.83INNAISVII78 pKa = 3.82
Molecular weight: 8.78 kDa
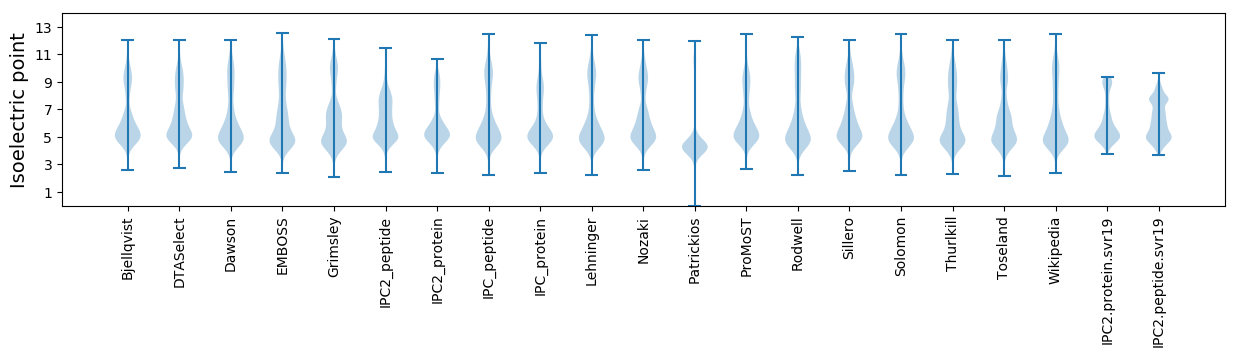
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T4E0F0|A0A2T4E0F0_9ALTE Acyl-coenzyme A dehydrogenase OS=Marinobacter sp. Z-F4-2 OX=2137199 GN=C9993_02600 PE=3 SV=1
MM1 pKa = 7.24SRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.74YY7 pKa = 9.74SPEE10 pKa = 3.78FKK12 pKa = 10.41RR13 pKa = 11.84EE14 pKa = 3.99AIALTRR20 pKa = 11.84QPGVSCRR27 pKa = 11.84QVALEE32 pKa = 4.24IGINPNLLSRR42 pKa = 11.84WRR44 pKa = 11.84RR45 pKa = 11.84EE46 pKa = 3.64ADD48 pKa = 3.35EE49 pKa = 4.4EE50 pKa = 4.06TDD52 pKa = 3.55KK53 pKa = 11.54AFKK56 pKa = 10.81GSGSPRR62 pKa = 11.84DD63 pKa = 3.6EE64 pKa = 3.44EE65 pKa = 4.31VARR68 pKa = 11.84LKK70 pKa = 10.99RR71 pKa = 11.84EE72 pKa = 3.69LARR75 pKa = 11.84VKK77 pKa = 10.55KK78 pKa = 10.29EE79 pKa = 3.49RR80 pKa = 11.84GFFARR85 pKa = 11.84SGSVLRR91 pKa = 11.84KK92 pKa = 9.88GVVLRR97 pKa = 11.84YY98 pKa = 9.18QAIQRR103 pKa = 11.84CRR105 pKa = 11.84NTFPIRR111 pKa = 11.84LMCRR115 pKa = 11.84CLKK118 pKa = 10.57VSASGYY124 pKa = 7.0YY125 pKa = 9.83AWVSRR130 pKa = 11.84PEE132 pKa = 4.12SARR135 pKa = 11.84SKK137 pKa = 11.05ANQHH141 pKa = 5.5LLSRR145 pKa = 11.84IRR147 pKa = 11.84EE148 pKa = 3.94IHH150 pKa = 7.05DD151 pKa = 4.18DD152 pKa = 3.73SGGMIGSPRR161 pKa = 11.84MHH163 pKa = 7.24EE164 pKa = 4.12DD165 pKa = 3.22LVAEE169 pKa = 4.6GASTSLNRR177 pKa = 11.84VARR180 pKa = 11.84LMAKK184 pKa = 10.01HH185 pKa = 6.92GIQGWPRR192 pKa = 11.84KK193 pKa = 9.58KK194 pKa = 10.05RR195 pKa = 11.84GRR197 pKa = 11.84LGRR200 pKa = 11.84QSYY203 pKa = 9.62RR204 pKa = 11.84PPGIRR209 pKa = 11.84NHH211 pKa = 6.25LEE213 pKa = 3.62RR214 pKa = 11.84DD215 pKa = 4.45FSALEE220 pKa = 4.44PDD222 pKa = 4.25TKK224 pKa = 10.22WVTDD228 pKa = 3.18ITEE231 pKa = 4.07IPTQEE236 pKa = 3.94GKK238 pKa = 10.91LFLCVVLDD246 pKa = 4.41LFSKK250 pKa = 10.8LIVGWSMHH258 pKa = 5.4HH259 pKa = 5.74RR260 pKa = 11.84QDD262 pKa = 2.6RR263 pKa = 11.84HH264 pKa = 3.58MVIRR268 pKa = 11.84AVEE271 pKa = 3.81MAVWQRR277 pKa = 11.84QGKK280 pKa = 9.2VPVILHH286 pKa = 6.05SDD288 pKa = 3.38RR289 pKa = 11.84GSQFTSGDD297 pKa = 3.41YY298 pKa = 10.36QRR300 pKa = 11.84YY301 pKa = 8.95LGSNRR306 pKa = 11.84LVSSMSAVGHH316 pKa = 6.43CGDD319 pKa = 3.77NAACEE324 pKa = 4.52GFFGMLKK331 pKa = 9.98RR332 pKa = 11.84EE333 pKa = 4.44RR334 pKa = 11.84IHH336 pKa = 5.73HH337 pKa = 5.51RR338 pKa = 11.84RR339 pKa = 11.84YY340 pKa = 8.89RR341 pKa = 11.84TQNEE345 pKa = 3.11ARR347 pKa = 11.84ADD349 pKa = 3.49VFDD352 pKa = 4.63YY353 pKa = 10.82IEE355 pKa = 4.7RR356 pKa = 11.84FHH358 pKa = 6.55NPRR361 pKa = 11.84MRR363 pKa = 11.84RR364 pKa = 11.84RR365 pKa = 11.84VAAQDD370 pKa = 3.12RR371 pKa = 11.84KK372 pKa = 10.57LFAVLKK378 pKa = 9.91PSVEE382 pKa = 4.31TGG384 pKa = 2.95
MM1 pKa = 7.24SRR3 pKa = 11.84RR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 9.74YY7 pKa = 9.74SPEE10 pKa = 3.78FKK12 pKa = 10.41RR13 pKa = 11.84EE14 pKa = 3.99AIALTRR20 pKa = 11.84QPGVSCRR27 pKa = 11.84QVALEE32 pKa = 4.24IGINPNLLSRR42 pKa = 11.84WRR44 pKa = 11.84RR45 pKa = 11.84EE46 pKa = 3.64ADD48 pKa = 3.35EE49 pKa = 4.4EE50 pKa = 4.06TDD52 pKa = 3.55KK53 pKa = 11.54AFKK56 pKa = 10.81GSGSPRR62 pKa = 11.84DD63 pKa = 3.6EE64 pKa = 3.44EE65 pKa = 4.31VARR68 pKa = 11.84LKK70 pKa = 10.99RR71 pKa = 11.84EE72 pKa = 3.69LARR75 pKa = 11.84VKK77 pKa = 10.55KK78 pKa = 10.29EE79 pKa = 3.49RR80 pKa = 11.84GFFARR85 pKa = 11.84SGSVLRR91 pKa = 11.84KK92 pKa = 9.88GVVLRR97 pKa = 11.84YY98 pKa = 9.18QAIQRR103 pKa = 11.84CRR105 pKa = 11.84NTFPIRR111 pKa = 11.84LMCRR115 pKa = 11.84CLKK118 pKa = 10.57VSASGYY124 pKa = 7.0YY125 pKa = 9.83AWVSRR130 pKa = 11.84PEE132 pKa = 4.12SARR135 pKa = 11.84SKK137 pKa = 11.05ANQHH141 pKa = 5.5LLSRR145 pKa = 11.84IRR147 pKa = 11.84EE148 pKa = 3.94IHH150 pKa = 7.05DD151 pKa = 4.18DD152 pKa = 3.73SGGMIGSPRR161 pKa = 11.84MHH163 pKa = 7.24EE164 pKa = 4.12DD165 pKa = 3.22LVAEE169 pKa = 4.6GASTSLNRR177 pKa = 11.84VARR180 pKa = 11.84LMAKK184 pKa = 10.01HH185 pKa = 6.92GIQGWPRR192 pKa = 11.84KK193 pKa = 9.58KK194 pKa = 10.05RR195 pKa = 11.84GRR197 pKa = 11.84LGRR200 pKa = 11.84QSYY203 pKa = 9.62RR204 pKa = 11.84PPGIRR209 pKa = 11.84NHH211 pKa = 6.25LEE213 pKa = 3.62RR214 pKa = 11.84DD215 pKa = 4.45FSALEE220 pKa = 4.44PDD222 pKa = 4.25TKK224 pKa = 10.22WVTDD228 pKa = 3.18ITEE231 pKa = 4.07IPTQEE236 pKa = 3.94GKK238 pKa = 10.91LFLCVVLDD246 pKa = 4.41LFSKK250 pKa = 10.8LIVGWSMHH258 pKa = 5.4HH259 pKa = 5.74RR260 pKa = 11.84QDD262 pKa = 2.6RR263 pKa = 11.84HH264 pKa = 3.58MVIRR268 pKa = 11.84AVEE271 pKa = 3.81MAVWQRR277 pKa = 11.84QGKK280 pKa = 9.2VPVILHH286 pKa = 6.05SDD288 pKa = 3.38RR289 pKa = 11.84GSQFTSGDD297 pKa = 3.41YY298 pKa = 10.36QRR300 pKa = 11.84YY301 pKa = 8.95LGSNRR306 pKa = 11.84LVSSMSAVGHH316 pKa = 6.43CGDD319 pKa = 3.77NAACEE324 pKa = 4.52GFFGMLKK331 pKa = 9.98RR332 pKa = 11.84EE333 pKa = 4.44RR334 pKa = 11.84IHH336 pKa = 5.73HH337 pKa = 5.51RR338 pKa = 11.84RR339 pKa = 11.84YY340 pKa = 8.89RR341 pKa = 11.84TQNEE345 pKa = 3.11ARR347 pKa = 11.84ADD349 pKa = 3.49VFDD352 pKa = 4.63YY353 pKa = 10.82IEE355 pKa = 4.7RR356 pKa = 11.84FHH358 pKa = 6.55NPRR361 pKa = 11.84MRR363 pKa = 11.84RR364 pKa = 11.84RR365 pKa = 11.84VAAQDD370 pKa = 3.12RR371 pKa = 11.84KK372 pKa = 10.57LFAVLKK378 pKa = 9.91PSVEE382 pKa = 4.31TGG384 pKa = 2.95
Molecular weight: 44.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
642497 |
20 |
1734 |
250.2 |
27.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.519 ± 0.062 | 0.96 ± 0.016 |
5.781 ± 0.037 | 6.833 ± 0.052 |
3.945 ± 0.035 | 7.523 ± 0.05 |
2.197 ± 0.029 | 5.298 ± 0.037 |
4.082 ± 0.055 | 10.558 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.475 ± 0.029 | 3.395 ± 0.033 |
4.511 ± 0.034 | 4.017 ± 0.043 |
6.477 ± 0.047 | 6.156 ± 0.039 |
5.064 ± 0.028 | 7.081 ± 0.047 |
1.427 ± 0.02 | 2.7 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
