
Candidatus Syntrophosphaera thermopropionivorans
Taxonomy: cellular organisms; Bacteria; FCB group; Candidatus Cloacimonetes; Candidatus Syntrophosphaera
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
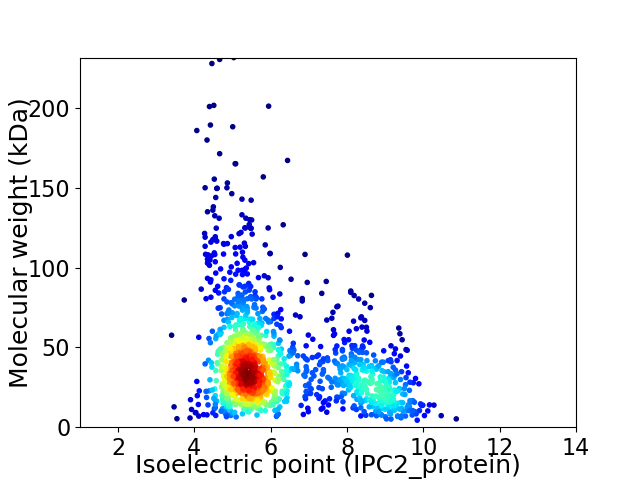
Virtual 2D-PAGE plot for 1369 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R5JQB6|A0A4R5JQB6_9BACT Glucose 1-dehydrogenase OS=Candidatus Syntrophosphaera thermopropionivorans OX=2593015 GN=E0946_04425 PE=4 SV=1
MM1 pKa = 7.42NKK3 pKa = 9.96YY4 pKa = 10.63LLLLLLLPVMIFAQPWQQDD23 pKa = 3.21NAIFNPSGIPSLPFSQPRR41 pKa = 11.84FVDD44 pKa = 3.94LDD46 pKa = 3.66ADD48 pKa = 3.59GDD50 pKa = 3.82MDD52 pKa = 5.0FFLGNTNSAPLYY64 pKa = 8.76ICNIGTATSPHH75 pKa = 5.94FVIGDD80 pKa = 3.59NLLANISFIDD90 pKa = 4.03AEE92 pKa = 4.41LAVCADD98 pKa = 3.62MNADD102 pKa = 3.69GKK104 pKa = 11.28LDD106 pKa = 5.62LITGGYY112 pKa = 7.81TGLHH116 pKa = 6.26LFLNTGTIQSPVYY129 pKa = 9.67TEE131 pKa = 3.57QLGYY135 pKa = 10.69FSTLEE140 pKa = 3.94VGNNPVPDD148 pKa = 3.73VADD151 pKa = 3.33VDD153 pKa = 4.14GDD155 pKa = 3.86GDD157 pKa = 4.37LDD159 pKa = 4.07LVVGLSEE166 pKa = 4.68DD167 pKa = 3.76GAVRR171 pKa = 11.84VYY173 pKa = 10.89FNTGTASNGIFQEE186 pKa = 4.43EE187 pKa = 4.69SMQLIGDD194 pKa = 4.04VGLYY198 pKa = 9.81AYY200 pKa = 9.47PVFCDD205 pKa = 4.05LDD207 pKa = 3.71NDD209 pKa = 3.99GDD211 pKa = 3.9QDD213 pKa = 3.88ILCGRR218 pKa = 11.84DD219 pKa = 2.93AHH221 pKa = 6.7GFVYY225 pKa = 10.2YY226 pKa = 10.43QNNGTISSPLWEE238 pKa = 4.57NNSALFSGLGNSTYY252 pKa = 10.19WNSPDD257 pKa = 5.67LVDD260 pKa = 5.92LDD262 pKa = 4.19GDD264 pKa = 4.04GDD266 pKa = 3.93FDD268 pKa = 6.91LIFGTADD275 pKa = 3.43GPLKK279 pKa = 10.82YY280 pKa = 10.07YY281 pKa = 10.73VNNGTPSSPQWQEE294 pKa = 3.39NTNLFGGVIDD304 pKa = 4.77AGGASSPVLFDD315 pKa = 3.97FDD317 pKa = 4.9GDD319 pKa = 3.68GDD321 pKa = 4.64LDD323 pKa = 5.05LICGTQLGDD332 pKa = 2.84IKK334 pKa = 10.66FYY336 pKa = 11.43KK337 pKa = 10.72NIGNPYY343 pKa = 8.97APAWQEE349 pKa = 3.65DD350 pKa = 3.39SSYY353 pKa = 10.82FSSIDD358 pKa = 3.18HH359 pKa = 6.81SIYY362 pKa = 10.79SAITLGDD369 pKa = 3.21VDD371 pKa = 6.04ADD373 pKa = 3.97GLPDD377 pKa = 5.54AIVGDD382 pKa = 4.2LSGNLYY388 pKa = 9.45FHH390 pKa = 7.47KK391 pKa = 10.41NTGTGFIQVNNVLPPISVGGWSVPRR416 pKa = 11.84LIDD419 pKa = 3.4MDD421 pKa = 4.43SDD423 pKa = 3.91GDD425 pKa = 3.78LDD427 pKa = 4.07LAVGNEE433 pKa = 3.88AGNIYY438 pKa = 10.35YY439 pKa = 9.27YY440 pKa = 10.76QNNGSPEE447 pKa = 4.1LPQWTLVSGFFGNIDD462 pKa = 3.58VGSDD466 pKa = 3.41CSPTFGDD473 pKa = 3.42LDD475 pKa = 3.97EE476 pKa = 5.03NGRR479 pKa = 11.84LDD481 pKa = 5.15LLAGDD486 pKa = 5.05LFGSLHH492 pKa = 6.79CYY494 pKa = 10.34LYY496 pKa = 10.82QNHH499 pKa = 5.7NWVNNTEE506 pKa = 4.07LFAGIEE512 pKa = 4.42TNQNAAPALADD523 pKa = 4.2LDD525 pKa = 4.19HH526 pKa = 7.53DD527 pKa = 4.84GDD529 pKa = 4.47LDD531 pKa = 4.81LVLGDD536 pKa = 3.76YY537 pKa = 11.23DD538 pKa = 4.0GTFKK542 pKa = 10.72FYY544 pKa = 11.17RR545 pKa = 11.84NLKK548 pKa = 9.51YY549 pKa = 10.79SPAVLNPPLNLLANTDD565 pKa = 3.84EE566 pKa = 4.64PVTISWEE573 pKa = 4.19EE574 pKa = 4.02PQGSTSPLEE583 pKa = 4.25HH584 pKa = 6.55YY585 pKa = 10.63NIYY588 pKa = 10.52LDD590 pKa = 3.96NEE592 pKa = 4.58FVDD595 pKa = 3.83STTFTFYY602 pKa = 11.46SFFNWDD608 pKa = 3.13FTVAHH613 pKa = 6.32IVKK616 pKa = 8.4VTAQYY621 pKa = 10.91VAGEE625 pKa = 4.27SVPAIIEE632 pKa = 4.1IPATPLNDD640 pKa = 3.47EE641 pKa = 4.28YY642 pKa = 11.56QSIGIISIYY651 pKa = 9.32PNPVTDD657 pKa = 3.27ITNINLTLKK666 pKa = 10.63SEE668 pKa = 4.56DD669 pKa = 3.45KK670 pKa = 10.64ATVSIYY676 pKa = 10.15NLKK679 pKa = 9.41GQQIIGWEE687 pKa = 4.31VVSKK691 pKa = 11.08GNHH694 pKa = 4.33NLIWNGKK701 pKa = 8.15DD702 pKa = 3.32KK703 pKa = 10.96QGKK706 pKa = 7.67PVANGIYY713 pKa = 9.89LLNVKK718 pKa = 8.72TPQNTITRR726 pKa = 11.84KK727 pKa = 9.58ISLIKK732 pKa = 10.61
MM1 pKa = 7.42NKK3 pKa = 9.96YY4 pKa = 10.63LLLLLLLPVMIFAQPWQQDD23 pKa = 3.21NAIFNPSGIPSLPFSQPRR41 pKa = 11.84FVDD44 pKa = 3.94LDD46 pKa = 3.66ADD48 pKa = 3.59GDD50 pKa = 3.82MDD52 pKa = 5.0FFLGNTNSAPLYY64 pKa = 8.76ICNIGTATSPHH75 pKa = 5.94FVIGDD80 pKa = 3.59NLLANISFIDD90 pKa = 4.03AEE92 pKa = 4.41LAVCADD98 pKa = 3.62MNADD102 pKa = 3.69GKK104 pKa = 11.28LDD106 pKa = 5.62LITGGYY112 pKa = 7.81TGLHH116 pKa = 6.26LFLNTGTIQSPVYY129 pKa = 9.67TEE131 pKa = 3.57QLGYY135 pKa = 10.69FSTLEE140 pKa = 3.94VGNNPVPDD148 pKa = 3.73VADD151 pKa = 3.33VDD153 pKa = 4.14GDD155 pKa = 3.86GDD157 pKa = 4.37LDD159 pKa = 4.07LVVGLSEE166 pKa = 4.68DD167 pKa = 3.76GAVRR171 pKa = 11.84VYY173 pKa = 10.89FNTGTASNGIFQEE186 pKa = 4.43EE187 pKa = 4.69SMQLIGDD194 pKa = 4.04VGLYY198 pKa = 9.81AYY200 pKa = 9.47PVFCDD205 pKa = 4.05LDD207 pKa = 3.71NDD209 pKa = 3.99GDD211 pKa = 3.9QDD213 pKa = 3.88ILCGRR218 pKa = 11.84DD219 pKa = 2.93AHH221 pKa = 6.7GFVYY225 pKa = 10.2YY226 pKa = 10.43QNNGTISSPLWEE238 pKa = 4.57NNSALFSGLGNSTYY252 pKa = 10.19WNSPDD257 pKa = 5.67LVDD260 pKa = 5.92LDD262 pKa = 4.19GDD264 pKa = 4.04GDD266 pKa = 3.93FDD268 pKa = 6.91LIFGTADD275 pKa = 3.43GPLKK279 pKa = 10.82YY280 pKa = 10.07YY281 pKa = 10.73VNNGTPSSPQWQEE294 pKa = 3.39NTNLFGGVIDD304 pKa = 4.77AGGASSPVLFDD315 pKa = 3.97FDD317 pKa = 4.9GDD319 pKa = 3.68GDD321 pKa = 4.64LDD323 pKa = 5.05LICGTQLGDD332 pKa = 2.84IKK334 pKa = 10.66FYY336 pKa = 11.43KK337 pKa = 10.72NIGNPYY343 pKa = 8.97APAWQEE349 pKa = 3.65DD350 pKa = 3.39SSYY353 pKa = 10.82FSSIDD358 pKa = 3.18HH359 pKa = 6.81SIYY362 pKa = 10.79SAITLGDD369 pKa = 3.21VDD371 pKa = 6.04ADD373 pKa = 3.97GLPDD377 pKa = 5.54AIVGDD382 pKa = 4.2LSGNLYY388 pKa = 9.45FHH390 pKa = 7.47KK391 pKa = 10.41NTGTGFIQVNNVLPPISVGGWSVPRR416 pKa = 11.84LIDD419 pKa = 3.4MDD421 pKa = 4.43SDD423 pKa = 3.91GDD425 pKa = 3.78LDD427 pKa = 4.07LAVGNEE433 pKa = 3.88AGNIYY438 pKa = 10.35YY439 pKa = 9.27YY440 pKa = 10.76QNNGSPEE447 pKa = 4.1LPQWTLVSGFFGNIDD462 pKa = 3.58VGSDD466 pKa = 3.41CSPTFGDD473 pKa = 3.42LDD475 pKa = 3.97EE476 pKa = 5.03NGRR479 pKa = 11.84LDD481 pKa = 5.15LLAGDD486 pKa = 5.05LFGSLHH492 pKa = 6.79CYY494 pKa = 10.34LYY496 pKa = 10.82QNHH499 pKa = 5.7NWVNNTEE506 pKa = 4.07LFAGIEE512 pKa = 4.42TNQNAAPALADD523 pKa = 4.2LDD525 pKa = 4.19HH526 pKa = 7.53DD527 pKa = 4.84GDD529 pKa = 4.47LDD531 pKa = 4.81LVLGDD536 pKa = 3.76YY537 pKa = 11.23DD538 pKa = 4.0GTFKK542 pKa = 10.72FYY544 pKa = 11.17RR545 pKa = 11.84NLKK548 pKa = 9.51YY549 pKa = 10.79SPAVLNPPLNLLANTDD565 pKa = 3.84EE566 pKa = 4.64PVTISWEE573 pKa = 4.19EE574 pKa = 4.02PQGSTSPLEE583 pKa = 4.25HH584 pKa = 6.55YY585 pKa = 10.63NIYY588 pKa = 10.52LDD590 pKa = 3.96NEE592 pKa = 4.58FVDD595 pKa = 3.83STTFTFYY602 pKa = 11.46SFFNWDD608 pKa = 3.13FTVAHH613 pKa = 6.32IVKK616 pKa = 8.4VTAQYY621 pKa = 10.91VAGEE625 pKa = 4.27SVPAIIEE632 pKa = 4.1IPATPLNDD640 pKa = 3.47EE641 pKa = 4.28YY642 pKa = 11.56QSIGIISIYY651 pKa = 9.32PNPVTDD657 pKa = 3.27ITNINLTLKK666 pKa = 10.63SEE668 pKa = 4.56DD669 pKa = 3.45KK670 pKa = 10.64ATVSIYY676 pKa = 10.15NLKK679 pKa = 9.41GQQIIGWEE687 pKa = 4.31VVSKK691 pKa = 11.08GNHH694 pKa = 4.33NLIWNGKK701 pKa = 8.15DD702 pKa = 3.32KK703 pKa = 10.96QGKK706 pKa = 7.67PVANGIYY713 pKa = 9.89LLNVKK718 pKa = 8.72TPQNTITRR726 pKa = 11.84KK727 pKa = 9.58ISLIKK732 pKa = 10.61
Molecular weight: 79.72 kDa
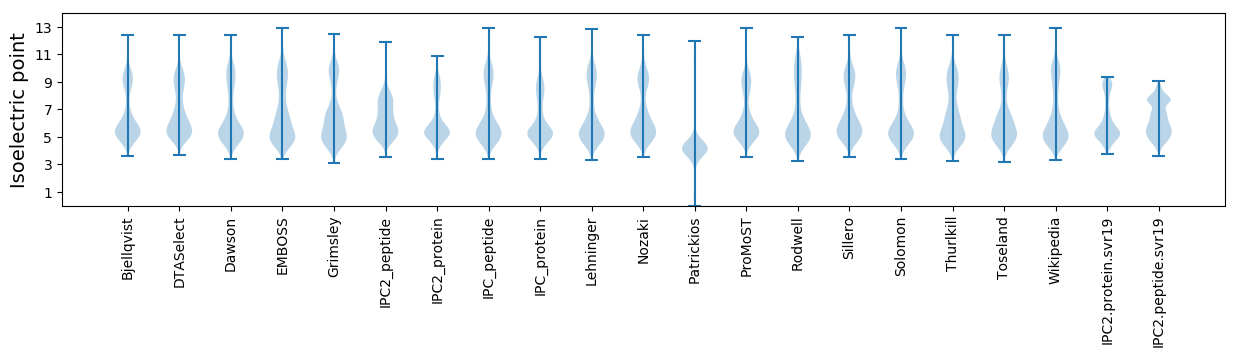
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R5JRB7|A0A4R5JRB7_9BACT Uncharacterized protein OS=Candidatus Syntrophosphaera thermopropionivorans OX=2593015 GN=E0946_02790 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.3QPSNRR10 pKa = 11.84SRR12 pKa = 11.84KK13 pKa = 7.31STHH16 pKa = 5.08GFRR19 pKa = 11.84SRR21 pKa = 11.84MATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 8.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 5.15TLCVV44 pKa = 3.33
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.3QPSNRR10 pKa = 11.84SRR12 pKa = 11.84KK13 pKa = 7.31STHH16 pKa = 5.08GFRR19 pKa = 11.84SRR21 pKa = 11.84MATRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 8.84VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 5.15TLCVV44 pKa = 3.33
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
524344 |
38 |
2085 |
383.0 |
43.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.423 ± 0.053 | 1.065 ± 0.021 |
5.272 ± 0.041 | 6.7 ± 0.078 |
4.392 ± 0.038 | 6.096 ± 0.057 |
1.844 ± 0.026 | 8.503 ± 0.056 |
6.338 ± 0.076 | 10.125 ± 0.072 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.264 ± 0.03 | 5.613 ± 0.07 |
4.467 ± 0.038 | 3.845 ± 0.037 |
3.996 ± 0.047 | 6.31 ± 0.059 |
5.38 ± 0.072 | 5.643 ± 0.051 |
1.164 ± 0.028 | 4.556 ± 0.058 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
