
Eubacterium plexicaudatum ASF492
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriaceae; Eubacterium; Eubacterium plexicaudatum
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
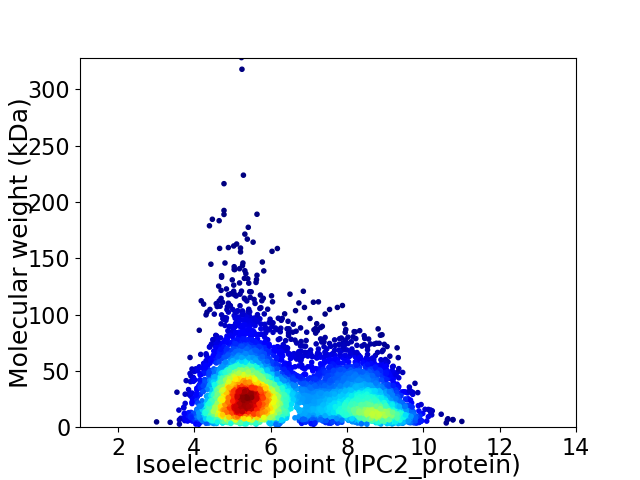
Virtual 2D-PAGE plot for 6154 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|N2A476|N2A476_9FIRM Uncharacterized protein (Fragment) OS=Eubacterium plexicaudatum ASF492 OX=1235802 GN=C823_04627 PE=4 SV=1
MM1 pKa = 7.5KK2 pKa = 10.4KK3 pKa = 10.32KK4 pKa = 10.22ILAVLLAGMMAFSMAACGSANDD26 pKa = 4.67GNADD30 pKa = 3.8SNKK33 pKa = 10.19EE34 pKa = 3.9NTDD37 pKa = 2.95NSGADD42 pKa = 3.3GSEE45 pKa = 4.04ADD47 pKa = 5.1DD48 pKa = 4.19SKK50 pKa = 11.99SDD52 pKa = 4.09DD53 pKa = 4.46GEE55 pKa = 4.42SSDD58 pKa = 5.83DD59 pKa = 3.97GSNDD63 pKa = 3.16DD64 pKa = 4.44SAPASDD70 pKa = 4.44MNIAMITDD78 pKa = 3.58SGDD81 pKa = 3.07ITDD84 pKa = 3.6QSFNQTTYY92 pKa = 8.47EE93 pKa = 4.04TSKK96 pKa = 10.97AWAEE100 pKa = 4.04ANNVPFTYY108 pKa = 10.25YY109 pKa = 10.72KK110 pKa = 10.04PDD112 pKa = 3.54SDD114 pKa = 4.46SDD116 pKa = 3.6EE117 pKa = 4.14ARR119 pKa = 11.84NASVDD124 pKa = 3.46QAVAGGANVLMMPGYY139 pKa = 9.11MFAASVVAKK148 pKa = 10.8AEE150 pKa = 4.03MYY152 pKa = 10.54PDD154 pKa = 3.46VKK156 pKa = 10.84FIALDD161 pKa = 3.48VGAGDD166 pKa = 4.39LLEE169 pKa = 5.14KK170 pKa = 10.8GVGEE174 pKa = 4.54GYY176 pKa = 10.67DD177 pKa = 4.08YY178 pKa = 11.58NPEE181 pKa = 3.97NYY183 pKa = 9.51DD184 pKa = 3.3VKK186 pKa = 10.76EE187 pKa = 4.13YY188 pKa = 10.55YY189 pKa = 10.2HH190 pKa = 7.3ADD192 pKa = 3.08NVYY195 pKa = 9.0CCTYY199 pKa = 10.33QEE201 pKa = 4.24EE202 pKa = 4.68LSGYY206 pKa = 7.19MAGYY210 pKa = 9.98AAVKK214 pKa = 10.23LGYY217 pKa = 9.73KK218 pKa = 10.17HH219 pKa = 6.66LGFLGGMSVPAVTRR233 pKa = 11.84FGYY236 pKa = 10.66GYY238 pKa = 10.48LQGADD243 pKa = 3.32AAAKK247 pKa = 9.43EE248 pKa = 4.03LGIEE252 pKa = 4.3GEE254 pKa = 4.29VEE256 pKa = 3.67AEE258 pKa = 4.33YY259 pKa = 11.24VCGGQFYY266 pKa = 11.29GDD268 pKa = 3.9ADD270 pKa = 3.24ITAAMDD276 pKa = 3.21TWYY279 pKa = 8.79GTKK282 pKa = 10.19GVEE285 pKa = 4.43VVFACGGGIFTSAVEE300 pKa = 4.21AAVKK304 pKa = 10.66ADD306 pKa = 3.36GKK308 pKa = 11.18VIGVDD313 pKa = 3.47SDD315 pKa = 3.34QSPIIDD321 pKa = 3.52QSKK324 pKa = 10.45EE325 pKa = 3.79GLTVTSAMKK334 pKa = 10.75GLATTVDD341 pKa = 3.56TVLTEE346 pKa = 4.02IQAGNWNNYY355 pKa = 8.16AGKK358 pKa = 9.89IEE360 pKa = 3.95NLGMVSEE367 pKa = 4.32NPEE370 pKa = 4.03EE371 pKa = 4.65NFVQLPISTTQWGDD385 pKa = 2.74GFTEE389 pKa = 4.05DD390 pKa = 5.33DD391 pKa = 3.65YY392 pKa = 11.37RR393 pKa = 11.84TLVKK397 pKa = 10.65AMYY400 pKa = 9.26NGEE403 pKa = 4.28VEE405 pKa = 4.17VSNDD409 pKa = 2.64IKK411 pKa = 11.13ALPDD415 pKa = 3.42SLSIKK420 pKa = 10.0VNDD423 pKa = 3.77YY424 pKa = 11.56GSIKK428 pKa = 10.63
MM1 pKa = 7.5KK2 pKa = 10.4KK3 pKa = 10.32KK4 pKa = 10.22ILAVLLAGMMAFSMAACGSANDD26 pKa = 4.67GNADD30 pKa = 3.8SNKK33 pKa = 10.19EE34 pKa = 3.9NTDD37 pKa = 2.95NSGADD42 pKa = 3.3GSEE45 pKa = 4.04ADD47 pKa = 5.1DD48 pKa = 4.19SKK50 pKa = 11.99SDD52 pKa = 4.09DD53 pKa = 4.46GEE55 pKa = 4.42SSDD58 pKa = 5.83DD59 pKa = 3.97GSNDD63 pKa = 3.16DD64 pKa = 4.44SAPASDD70 pKa = 4.44MNIAMITDD78 pKa = 3.58SGDD81 pKa = 3.07ITDD84 pKa = 3.6QSFNQTTYY92 pKa = 8.47EE93 pKa = 4.04TSKK96 pKa = 10.97AWAEE100 pKa = 4.04ANNVPFTYY108 pKa = 10.25YY109 pKa = 10.72KK110 pKa = 10.04PDD112 pKa = 3.54SDD114 pKa = 4.46SDD116 pKa = 3.6EE117 pKa = 4.14ARR119 pKa = 11.84NASVDD124 pKa = 3.46QAVAGGANVLMMPGYY139 pKa = 9.11MFAASVVAKK148 pKa = 10.8AEE150 pKa = 4.03MYY152 pKa = 10.54PDD154 pKa = 3.46VKK156 pKa = 10.84FIALDD161 pKa = 3.48VGAGDD166 pKa = 4.39LLEE169 pKa = 5.14KK170 pKa = 10.8GVGEE174 pKa = 4.54GYY176 pKa = 10.67DD177 pKa = 4.08YY178 pKa = 11.58NPEE181 pKa = 3.97NYY183 pKa = 9.51DD184 pKa = 3.3VKK186 pKa = 10.76EE187 pKa = 4.13YY188 pKa = 10.55YY189 pKa = 10.2HH190 pKa = 7.3ADD192 pKa = 3.08NVYY195 pKa = 9.0CCTYY199 pKa = 10.33QEE201 pKa = 4.24EE202 pKa = 4.68LSGYY206 pKa = 7.19MAGYY210 pKa = 9.98AAVKK214 pKa = 10.23LGYY217 pKa = 9.73KK218 pKa = 10.17HH219 pKa = 6.66LGFLGGMSVPAVTRR233 pKa = 11.84FGYY236 pKa = 10.66GYY238 pKa = 10.48LQGADD243 pKa = 3.32AAAKK247 pKa = 9.43EE248 pKa = 4.03LGIEE252 pKa = 4.3GEE254 pKa = 4.29VEE256 pKa = 3.67AEE258 pKa = 4.33YY259 pKa = 11.24VCGGQFYY266 pKa = 11.29GDD268 pKa = 3.9ADD270 pKa = 3.24ITAAMDD276 pKa = 3.21TWYY279 pKa = 8.79GTKK282 pKa = 10.19GVEE285 pKa = 4.43VVFACGGGIFTSAVEE300 pKa = 4.21AAVKK304 pKa = 10.66ADD306 pKa = 3.36GKK308 pKa = 11.18VIGVDD313 pKa = 3.47SDD315 pKa = 3.34QSPIIDD321 pKa = 3.52QSKK324 pKa = 10.45EE325 pKa = 3.79GLTVTSAMKK334 pKa = 10.75GLATTVDD341 pKa = 3.56TVLTEE346 pKa = 4.02IQAGNWNNYY355 pKa = 8.16AGKK358 pKa = 9.89IEE360 pKa = 3.95NLGMVSEE367 pKa = 4.32NPEE370 pKa = 4.03EE371 pKa = 4.65NFVQLPISTTQWGDD385 pKa = 2.74GFTEE389 pKa = 4.05DD390 pKa = 5.33DD391 pKa = 3.65YY392 pKa = 11.37RR393 pKa = 11.84TLVKK397 pKa = 10.65AMYY400 pKa = 9.26NGEE403 pKa = 4.28VEE405 pKa = 4.17VSNDD409 pKa = 2.64IKK411 pKa = 11.13ALPDD415 pKa = 3.42SLSIKK420 pKa = 10.0VNDD423 pKa = 3.77YY424 pKa = 11.56GSIKK428 pKa = 10.63
Molecular weight: 45.42 kDa
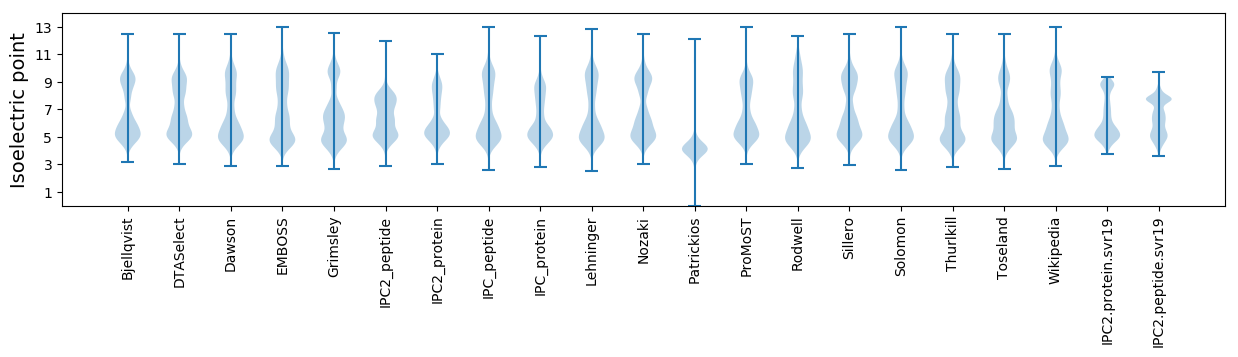
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|N2AY61|N2AY61_9FIRM Uncharacterized protein OS=Eubacterium plexicaudatum ASF492 OX=1235802 GN=C823_01138 PE=4 SV=1
MM1 pKa = 7.66KK2 pKa = 8.5MTYY5 pKa = 9.73QPNKK9 pKa = 7.37RR10 pKa = 11.84HH11 pKa = 5.91RR12 pKa = 11.84SKK14 pKa = 10.83VHH16 pKa = 5.68GFRR19 pKa = 11.84KK20 pKa = 9.97RR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.62VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
MM1 pKa = 7.66KK2 pKa = 8.5MTYY5 pKa = 9.73QPNKK9 pKa = 7.37RR10 pKa = 11.84HH11 pKa = 5.91RR12 pKa = 11.84SKK14 pKa = 10.83VHH16 pKa = 5.68GFRR19 pKa = 11.84KK20 pKa = 9.97RR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.62VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1750650 |
20 |
2922 |
284.5 |
32.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.352 ± 0.039 | 1.649 ± 0.015 |
5.662 ± 0.022 | 7.485 ± 0.033 |
4.146 ± 0.026 | 6.537 ± 0.033 |
1.916 ± 0.013 | 7.204 ± 0.029 |
6.977 ± 0.034 | 8.874 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.181 ± 0.019 | 4.437 ± 0.025 |
3.094 ± 0.019 | 3.809 ± 0.023 |
4.885 ± 0.027 | 5.78 ± 0.025 |
5.197 ± 0.031 | 6.403 ± 0.025 |
0.99 ± 0.011 | 4.423 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
