
Blastococcus sp. DSM 46838
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Blastococcus; unclassified Blastococcus
Average proteome isoelectric point is 6.12
Get precalculated fractions of proteins
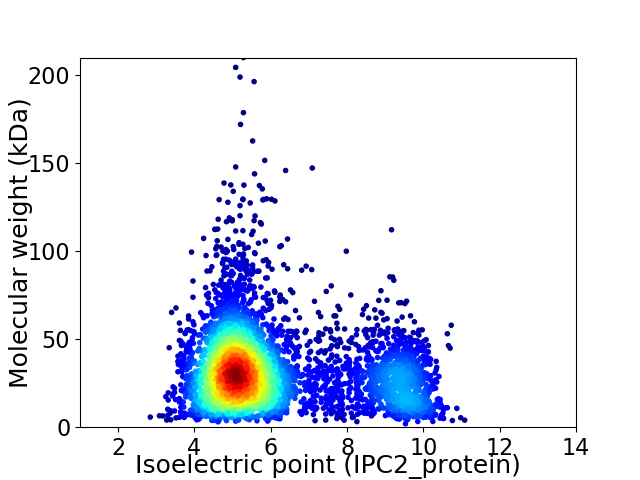
Virtual 2D-PAGE plot for 4797 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I2DZY8|A0A1I2DZY8_9ACTN Cation-transporting ATPase E OS=Blastococcus sp. DSM 46838 OX=1798228 GN=SAMN05216574_106186 PE=4 SV=1
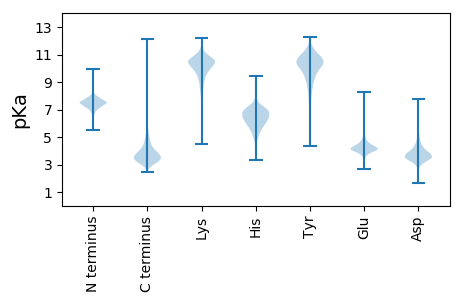
MM1 pKa = 7.15RR2 pKa = 11.84TRR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 10.46GSTGAAALAVALLAACSSGTTDD28 pKa = 4.26ASGGSDD34 pKa = 3.01GGASADD40 pKa = 3.62GPLAEE45 pKa = 4.78VVADD49 pKa = 3.59VDD51 pKa = 3.59AAAQARR57 pKa = 11.84DD58 pKa = 3.64TFAVPSEE65 pKa = 4.07PVDD68 pKa = 3.49VSGWEE73 pKa = 3.98GRR75 pKa = 11.84TVYY78 pKa = 9.85YY79 pKa = 10.73VPITAQISVFQVYY92 pKa = 9.87GEE94 pKa = 4.61KK95 pKa = 9.73IGEE98 pKa = 4.06ALEE101 pKa = 4.38TVGADD106 pKa = 4.38LQICDD111 pKa = 4.05GGSNPSQISGCIDD124 pKa = 3.28QAVGASAAAIITDD137 pKa = 3.5NVPYY141 pKa = 10.95GMAANALDD149 pKa = 4.07GARR152 pKa = 11.84SAGIPVLIANQLPDD166 pKa = 4.01PAHH169 pKa = 6.65PADD172 pKa = 3.74EE173 pKa = 4.14SLAYY177 pKa = 10.57VEE179 pKa = 5.81GPATEE184 pKa = 4.86MITSVADD191 pKa = 3.39WVVADD196 pKa = 4.31SDD198 pKa = 4.33GEE200 pKa = 4.34ATVVLQKK207 pKa = 9.83VTDD210 pKa = 3.96NPSTIAWAEE219 pKa = 3.85AAEE222 pKa = 3.91QRR224 pKa = 11.84IDD226 pKa = 4.17EE227 pKa = 4.62RR228 pKa = 11.84CPDD231 pKa = 3.49CTVVVNEE238 pKa = 4.19VSASNFPLIPSSTSSALLANPDD260 pKa = 3.16ARR262 pKa = 11.84YY263 pKa = 9.9VLAEE267 pKa = 4.26FEE269 pKa = 5.3HH270 pKa = 6.64FVQPTLGGIQQSPNAADD287 pKa = 3.31IKK289 pKa = 10.46IVASAATLSGLQMLADD305 pKa = 5.03GAQLHH310 pKa = 6.98ADD312 pKa = 3.67AGQNFAFQGWATADD326 pKa = 3.2ALFRR330 pKa = 11.84LVAGQEE336 pKa = 3.88IPEE339 pKa = 3.99YY340 pKa = 10.23DD341 pKa = 3.34YY342 pKa = 11.78SFRR345 pKa = 11.84LFTQDD350 pKa = 3.66NVADD354 pKa = 4.91LDD356 pKa = 4.69LSDD359 pKa = 4.85EE360 pKa = 4.51GQASGSWYY368 pKa = 10.58GPTDD372 pKa = 3.59YY373 pKa = 11.38ADD375 pKa = 3.45EE376 pKa = 4.66FAALWVEE383 pKa = 4.55GG384 pKa = 3.93
MM1 pKa = 7.15RR2 pKa = 11.84TRR4 pKa = 11.84RR5 pKa = 11.84YY6 pKa = 10.46GSTGAAALAVALLAACSSGTTDD28 pKa = 4.26ASGGSDD34 pKa = 3.01GGASADD40 pKa = 3.62GPLAEE45 pKa = 4.78VVADD49 pKa = 3.59VDD51 pKa = 3.59AAAQARR57 pKa = 11.84DD58 pKa = 3.64TFAVPSEE65 pKa = 4.07PVDD68 pKa = 3.49VSGWEE73 pKa = 3.98GRR75 pKa = 11.84TVYY78 pKa = 9.85YY79 pKa = 10.73VPITAQISVFQVYY92 pKa = 9.87GEE94 pKa = 4.61KK95 pKa = 9.73IGEE98 pKa = 4.06ALEE101 pKa = 4.38TVGADD106 pKa = 4.38LQICDD111 pKa = 4.05GGSNPSQISGCIDD124 pKa = 3.28QAVGASAAAIITDD137 pKa = 3.5NVPYY141 pKa = 10.95GMAANALDD149 pKa = 4.07GARR152 pKa = 11.84SAGIPVLIANQLPDD166 pKa = 4.01PAHH169 pKa = 6.65PADD172 pKa = 3.74EE173 pKa = 4.14SLAYY177 pKa = 10.57VEE179 pKa = 5.81GPATEE184 pKa = 4.86MITSVADD191 pKa = 3.39WVVADD196 pKa = 4.31SDD198 pKa = 4.33GEE200 pKa = 4.34ATVVLQKK207 pKa = 9.83VTDD210 pKa = 3.96NPSTIAWAEE219 pKa = 3.85AAEE222 pKa = 3.91QRR224 pKa = 11.84IDD226 pKa = 4.17EE227 pKa = 4.62RR228 pKa = 11.84CPDD231 pKa = 3.49CTVVVNEE238 pKa = 4.19VSASNFPLIPSSTSSALLANPDD260 pKa = 3.16ARR262 pKa = 11.84YY263 pKa = 9.9VLAEE267 pKa = 4.26FEE269 pKa = 5.3HH270 pKa = 6.64FVQPTLGGIQQSPNAADD287 pKa = 3.31IKK289 pKa = 10.46IVASAATLSGLQMLADD305 pKa = 5.03GAQLHH310 pKa = 6.98ADD312 pKa = 3.67AGQNFAFQGWATADD326 pKa = 3.2ALFRR330 pKa = 11.84LVAGQEE336 pKa = 3.88IPEE339 pKa = 3.99YY340 pKa = 10.23DD341 pKa = 3.34YY342 pKa = 11.78SFRR345 pKa = 11.84LFTQDD350 pKa = 3.66NVADD354 pKa = 4.91LDD356 pKa = 4.69LSDD359 pKa = 4.85EE360 pKa = 4.51GQASGSWYY368 pKa = 10.58GPTDD372 pKa = 3.59YY373 pKa = 11.38ADD375 pKa = 3.45EE376 pKa = 4.66FAALWVEE383 pKa = 4.55GG384 pKa = 3.93
Molecular weight: 39.68 kDa
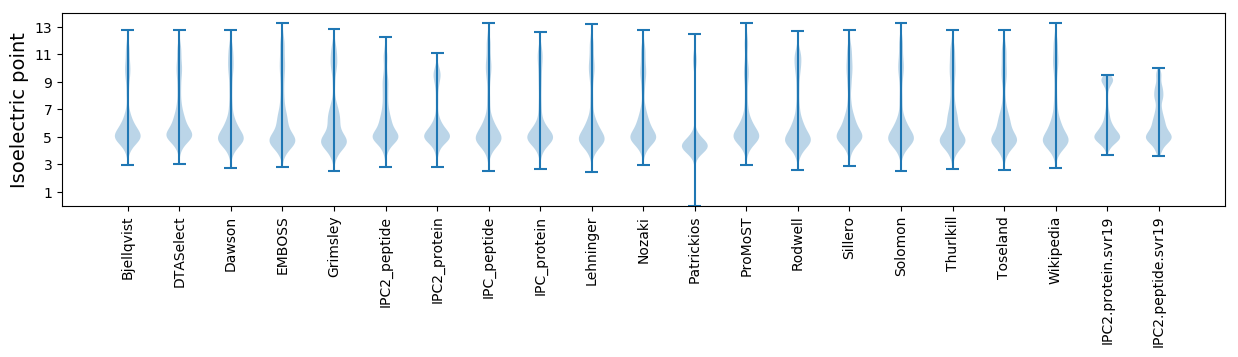
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I1VWJ0|A0A1I1VWJ0_9ACTN o-succinylbenzoate synthase OS=Blastococcus sp. DSM 46838 OX=1798228 GN=menC PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.99KK16 pKa = 9.77HH17 pKa = 5.81RR18 pKa = 11.84KK19 pKa = 8.51LLKK22 pKa = 8.15KK23 pKa = 9.25TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.81KK32 pKa = 9.63
Molecular weight: 4.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1512024 |
19 |
1967 |
315.2 |
33.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.303 ± 0.062 | 0.687 ± 0.009 |
6.276 ± 0.03 | 5.626 ± 0.033 |
2.589 ± 0.02 | 9.697 ± 0.035 |
2.013 ± 0.014 | 2.978 ± 0.025 |
1.381 ± 0.022 | 10.699 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.663 ± 0.014 | 1.444 ± 0.018 |
6.234 ± 0.031 | 2.701 ± 0.017 |
8.192 ± 0.036 | 4.846 ± 0.023 |
5.864 ± 0.024 | 9.605 ± 0.038 |
1.47 ± 0.015 | 1.734 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
