
Antarctobacter heliothermus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Antarctobacter
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
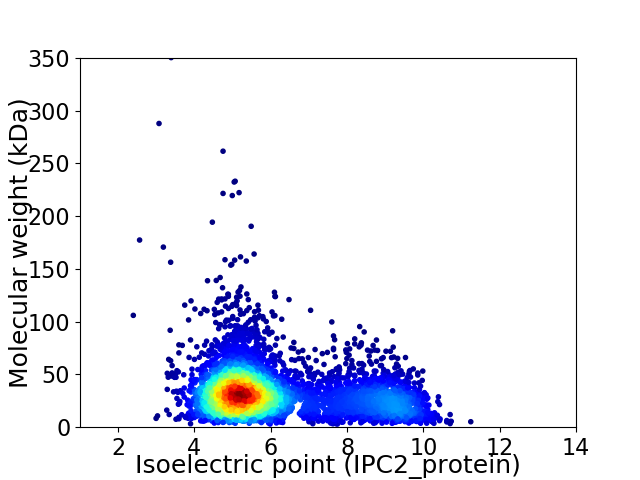
Virtual 2D-PAGE plot for 4962 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
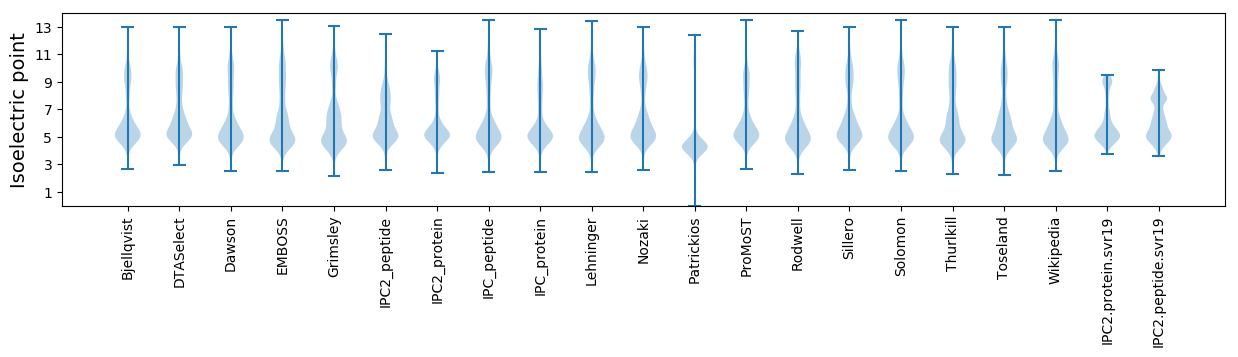
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A222E7N2|A0A222E7N2_9RHOB Deoxyguanosinetriphosphate triphosphohydrolase-like protein OS=Antarctobacter heliothermus OX=74033 GN=dgt PE=3 SV=1
MM1 pKa = 7.78CFLCQSLNPRR11 pKa = 11.84TDD13 pKa = 3.36DD14 pKa = 3.92YY15 pKa = 11.41EE16 pKa = 4.37FHH18 pKa = 6.99GLTGPTAAATVGDD31 pKa = 4.35RR32 pKa = 11.84SSAPASSKK40 pKa = 10.29PVYY43 pKa = 10.78SLDD46 pKa = 3.61QVATQLTHH54 pKa = 7.24GYY56 pKa = 7.12WQSGGGDD63 pKa = 2.69WRR65 pKa = 11.84AFDD68 pKa = 3.8VQAGDD73 pKa = 3.69TLTVNLDD80 pKa = 3.25GLDD83 pKa = 3.77PVGQAAAVAALAAWTAVSGLQFMTTSGDD111 pKa = 3.31ADD113 pKa = 3.43ITFQDD118 pKa = 3.65IDD120 pKa = 3.7GGAYY124 pKa = 9.98SYY126 pKa = 11.35SAVYY130 pKa = 9.12TGWNEE135 pKa = 3.26IAYY138 pKa = 10.25SVVNVHH144 pKa = 7.36ASWQGNADD152 pKa = 3.43YY153 pKa = 10.88YY154 pKa = 10.34YY155 pKa = 9.23QTFIHH160 pKa = 6.87EE161 pKa = 4.39IGHH164 pKa = 6.36ALGLGHH170 pKa = 7.13GGNYY174 pKa = 9.87NGSADD179 pKa = 4.05FDD181 pKa = 4.38SQAHH185 pKa = 5.16YY186 pKa = 11.47ANDD189 pKa = 3.25SWQMSIMSYY198 pKa = 9.67FAQWEE203 pKa = 4.36NPNVDD208 pKa = 3.1ASGMYY213 pKa = 10.58LNSPQMADD221 pKa = 2.72ILAIQNLYY229 pKa = 8.15GTPDD233 pKa = 3.39NVHH236 pKa = 6.24VGNSVYY242 pKa = 11.04GDD244 pKa = 3.74NSNLDD249 pKa = 3.49ATGMDD254 pKa = 4.33LEE256 pKa = 4.66RR257 pKa = 11.84GRR259 pKa = 11.84AVTIFDD265 pKa = 3.72SSGIDD270 pKa = 4.3LIDD273 pKa = 4.12LGSRR277 pKa = 11.84AYY279 pKa = 10.27NQYY282 pKa = 11.17LSLIDD287 pKa = 3.95ATWSNLDD294 pKa = 3.56GYY296 pKa = 10.75VGNFGIARR304 pKa = 11.84GAVIEE309 pKa = 3.99NAITGAGNDD318 pKa = 3.44HH319 pKa = 5.65VTGNTAANDD328 pKa = 3.35IQTGDD333 pKa = 3.52GADD336 pKa = 3.44TVQAGAGDD344 pKa = 3.79DD345 pKa = 4.33TIDD348 pKa = 3.75GGAGEE353 pKa = 4.64DD354 pKa = 3.58TVIFTGASEE363 pKa = 4.84GYY365 pKa = 10.57DD366 pKa = 4.16LIWDD370 pKa = 4.06GALRR374 pKa = 11.84ITDD377 pKa = 3.6TDD379 pKa = 3.87LTDD382 pKa = 4.59GDD384 pKa = 5.15DD385 pKa = 4.11GTDD388 pKa = 3.01VLTDD392 pKa = 3.42VEE394 pKa = 4.78TLSFTDD400 pKa = 4.27GAFGTIQSDD409 pKa = 3.97GAITKK414 pKa = 9.56VRR416 pKa = 11.84LYY418 pKa = 9.67KK419 pKa = 10.33TGSSLTEE426 pKa = 3.58EE427 pKa = 4.17TLEE430 pKa = 3.91VDD432 pKa = 4.22VANSHH437 pKa = 6.45DD438 pKa = 3.63WQSIARR444 pKa = 11.84SFTDD448 pKa = 3.14TGQWASQTNTYY459 pKa = 11.38DD460 pKa = 3.01NGRR463 pKa = 11.84VLQISFADD471 pKa = 3.93GLRR474 pKa = 11.84AQSTMTDD481 pKa = 3.26TANVYY486 pKa = 9.94SWASYY491 pKa = 9.02TDD493 pKa = 3.41IYY495 pKa = 11.22DD496 pKa = 3.41EE497 pKa = 4.25TGARR501 pKa = 11.84IINSYY506 pKa = 8.3TWDD509 pKa = 3.44SGKK512 pKa = 9.51MVDD515 pKa = 4.33VYY517 pKa = 11.33YY518 pKa = 10.62DD519 pKa = 3.48SEE521 pKa = 4.27GARR524 pKa = 11.84IGSLVIDD531 pKa = 4.82GGDD534 pKa = 3.17QFVWHH539 pKa = 6.92SIARR543 pKa = 11.84SYY545 pKa = 11.73DD546 pKa = 3.3STGTLTEE553 pKa = 4.05QNNLFDD559 pKa = 5.76DD560 pKa = 4.76GSEE563 pKa = 4.02QVISYY568 pKa = 9.73TDD570 pKa = 3.68GVRR573 pKa = 11.84SGISMTDD580 pKa = 3.41GADD583 pKa = 3.21KK584 pKa = 10.9AIWSSYY590 pKa = 8.26TDD592 pKa = 3.25TYY594 pKa = 11.25DD595 pKa = 3.09TATGAKK601 pKa = 9.28VSRR604 pKa = 11.84LMIYY608 pKa = 10.6DD609 pKa = 4.1DD610 pKa = 3.64GRR612 pKa = 11.84EE613 pKa = 3.98IEE615 pKa = 4.07TGYY618 pKa = 11.33DD619 pKa = 3.05NGQMVSQTITDD630 pKa = 3.87GADD633 pKa = 3.01DD634 pKa = 4.58FVWHH638 pKa = 5.88STTRR642 pKa = 11.84NYY644 pKa = 11.04DD645 pKa = 3.37SEE647 pKa = 4.86GMLDD651 pKa = 3.94SQVDD655 pKa = 3.89TYY657 pKa = 11.94DD658 pKa = 3.59DD659 pKa = 3.78GRR661 pKa = 11.84VMEE664 pKa = 4.89IEE666 pKa = 4.25FEE668 pKa = 4.09NGLRR672 pKa = 11.84SSSIQTDD679 pKa = 3.45VADD682 pKa = 4.7AYY684 pKa = 10.23AWASATEE691 pKa = 4.43TYY693 pKa = 10.74DD694 pKa = 4.49DD695 pKa = 4.12SGTLIEE701 pKa = 6.08RR702 pKa = 11.84VTHH705 pKa = 6.46WDD707 pKa = 3.75DD708 pKa = 3.73GSEE711 pKa = 4.2SAVTYY716 pKa = 10.55GDD718 pKa = 4.16SIAA721 pKa = 4.57
MM1 pKa = 7.78CFLCQSLNPRR11 pKa = 11.84TDD13 pKa = 3.36DD14 pKa = 3.92YY15 pKa = 11.41EE16 pKa = 4.37FHH18 pKa = 6.99GLTGPTAAATVGDD31 pKa = 4.35RR32 pKa = 11.84SSAPASSKK40 pKa = 10.29PVYY43 pKa = 10.78SLDD46 pKa = 3.61QVATQLTHH54 pKa = 7.24GYY56 pKa = 7.12WQSGGGDD63 pKa = 2.69WRR65 pKa = 11.84AFDD68 pKa = 3.8VQAGDD73 pKa = 3.69TLTVNLDD80 pKa = 3.25GLDD83 pKa = 3.77PVGQAAAVAALAAWTAVSGLQFMTTSGDD111 pKa = 3.31ADD113 pKa = 3.43ITFQDD118 pKa = 3.65IDD120 pKa = 3.7GGAYY124 pKa = 9.98SYY126 pKa = 11.35SAVYY130 pKa = 9.12TGWNEE135 pKa = 3.26IAYY138 pKa = 10.25SVVNVHH144 pKa = 7.36ASWQGNADD152 pKa = 3.43YY153 pKa = 10.88YY154 pKa = 10.34YY155 pKa = 9.23QTFIHH160 pKa = 6.87EE161 pKa = 4.39IGHH164 pKa = 6.36ALGLGHH170 pKa = 7.13GGNYY174 pKa = 9.87NGSADD179 pKa = 4.05FDD181 pKa = 4.38SQAHH185 pKa = 5.16YY186 pKa = 11.47ANDD189 pKa = 3.25SWQMSIMSYY198 pKa = 9.67FAQWEE203 pKa = 4.36NPNVDD208 pKa = 3.1ASGMYY213 pKa = 10.58LNSPQMADD221 pKa = 2.72ILAIQNLYY229 pKa = 8.15GTPDD233 pKa = 3.39NVHH236 pKa = 6.24VGNSVYY242 pKa = 11.04GDD244 pKa = 3.74NSNLDD249 pKa = 3.49ATGMDD254 pKa = 4.33LEE256 pKa = 4.66RR257 pKa = 11.84GRR259 pKa = 11.84AVTIFDD265 pKa = 3.72SSGIDD270 pKa = 4.3LIDD273 pKa = 4.12LGSRR277 pKa = 11.84AYY279 pKa = 10.27NQYY282 pKa = 11.17LSLIDD287 pKa = 3.95ATWSNLDD294 pKa = 3.56GYY296 pKa = 10.75VGNFGIARR304 pKa = 11.84GAVIEE309 pKa = 3.99NAITGAGNDD318 pKa = 3.44HH319 pKa = 5.65VTGNTAANDD328 pKa = 3.35IQTGDD333 pKa = 3.52GADD336 pKa = 3.44TVQAGAGDD344 pKa = 3.79DD345 pKa = 4.33TIDD348 pKa = 3.75GGAGEE353 pKa = 4.64DD354 pKa = 3.58TVIFTGASEE363 pKa = 4.84GYY365 pKa = 10.57DD366 pKa = 4.16LIWDD370 pKa = 4.06GALRR374 pKa = 11.84ITDD377 pKa = 3.6TDD379 pKa = 3.87LTDD382 pKa = 4.59GDD384 pKa = 5.15DD385 pKa = 4.11GTDD388 pKa = 3.01VLTDD392 pKa = 3.42VEE394 pKa = 4.78TLSFTDD400 pKa = 4.27GAFGTIQSDD409 pKa = 3.97GAITKK414 pKa = 9.56VRR416 pKa = 11.84LYY418 pKa = 9.67KK419 pKa = 10.33TGSSLTEE426 pKa = 3.58EE427 pKa = 4.17TLEE430 pKa = 3.91VDD432 pKa = 4.22VANSHH437 pKa = 6.45DD438 pKa = 3.63WQSIARR444 pKa = 11.84SFTDD448 pKa = 3.14TGQWASQTNTYY459 pKa = 11.38DD460 pKa = 3.01NGRR463 pKa = 11.84VLQISFADD471 pKa = 3.93GLRR474 pKa = 11.84AQSTMTDD481 pKa = 3.26TANVYY486 pKa = 9.94SWASYY491 pKa = 9.02TDD493 pKa = 3.41IYY495 pKa = 11.22DD496 pKa = 3.41EE497 pKa = 4.25TGARR501 pKa = 11.84IINSYY506 pKa = 8.3TWDD509 pKa = 3.44SGKK512 pKa = 9.51MVDD515 pKa = 4.33VYY517 pKa = 11.33YY518 pKa = 10.62DD519 pKa = 3.48SEE521 pKa = 4.27GARR524 pKa = 11.84IGSLVIDD531 pKa = 4.82GGDD534 pKa = 3.17QFVWHH539 pKa = 6.92SIARR543 pKa = 11.84SYY545 pKa = 11.73DD546 pKa = 3.3STGTLTEE553 pKa = 4.05QNNLFDD559 pKa = 5.76DD560 pKa = 4.76GSEE563 pKa = 4.02QVISYY568 pKa = 9.73TDD570 pKa = 3.68GVRR573 pKa = 11.84SGISMTDD580 pKa = 3.41GADD583 pKa = 3.21KK584 pKa = 10.9AIWSSYY590 pKa = 8.26TDD592 pKa = 3.25TYY594 pKa = 11.25DD595 pKa = 3.09TATGAKK601 pKa = 9.28VSRR604 pKa = 11.84LMIYY608 pKa = 10.6DD609 pKa = 4.1DD610 pKa = 3.64GRR612 pKa = 11.84EE613 pKa = 3.98IEE615 pKa = 4.07TGYY618 pKa = 11.33DD619 pKa = 3.05NGQMVSQTITDD630 pKa = 3.87GADD633 pKa = 3.01DD634 pKa = 4.58FVWHH638 pKa = 5.88STTRR642 pKa = 11.84NYY644 pKa = 11.04DD645 pKa = 3.37SEE647 pKa = 4.86GMLDD651 pKa = 3.94SQVDD655 pKa = 3.89TYY657 pKa = 11.94DD658 pKa = 3.59DD659 pKa = 3.78GRR661 pKa = 11.84VMEE664 pKa = 4.89IEE666 pKa = 4.25FEE668 pKa = 4.09NGLRR672 pKa = 11.84SSSIQTDD679 pKa = 3.45VADD682 pKa = 4.7AYY684 pKa = 10.23AWASATEE691 pKa = 4.43TYY693 pKa = 10.74DD694 pKa = 4.49DD695 pKa = 4.12SGTLIEE701 pKa = 6.08RR702 pKa = 11.84VTHH705 pKa = 6.46WDD707 pKa = 3.75DD708 pKa = 3.73GSEE711 pKa = 4.2SAVTYY716 pKa = 10.55GDD718 pKa = 4.16SIAA721 pKa = 4.57
Molecular weight: 77.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A222E225|A0A222E225_9RHOB D-alanyl-D-alanine carboxypeptidase DacF OS=Antarctobacter heliothermus OX=74033 GN=dacF PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.5ILNARR34 pKa = 11.84RR35 pKa = 11.84LRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.5ILNARR34 pKa = 11.84RR35 pKa = 11.84LRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1553299 |
29 |
3565 |
313.0 |
33.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.146 ± 0.048 | 0.944 ± 0.011 |
6.335 ± 0.044 | 5.591 ± 0.036 |
3.675 ± 0.02 | 8.804 ± 0.051 |
2.08 ± 0.02 | 5.034 ± 0.029 |
2.981 ± 0.032 | 10.128 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.791 ± 0.019 | 2.55 ± 0.022 |
5.202 ± 0.031 | 3.195 ± 0.02 |
6.722 ± 0.041 | 5.109 ± 0.023 |
5.684 ± 0.035 | 7.418 ± 0.032 |
1.409 ± 0.016 | 2.204 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
