
Wuhan heteroptera virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.9
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
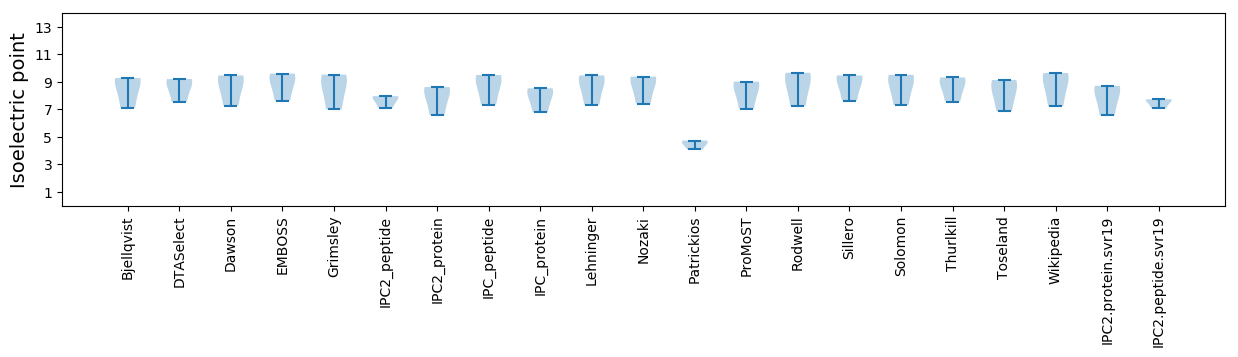
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A1L3KEV8|A0A1L3KEV8_9VIRU Capsid protein OS=Wuhan heteroptera virus 2 OX=1923702 PE=3 SV=1
MM1 pKa = 7.78DD2 pKa = 4.59FDD4 pKa = 4.08VRR6 pKa = 11.84VGRR9 pKa = 11.84ALKK12 pKa = 9.9EE13 pKa = 4.07CQPDD17 pKa = 3.7SVTGLGYY24 pKa = 10.23FRR26 pKa = 11.84NYY28 pKa = 7.2QTIGDD33 pKa = 4.21AFGHH37 pKa = 5.77EE38 pKa = 4.45CGFYY42 pKa = 10.61TNAHH46 pKa = 6.42NIAILVNIVRR56 pKa = 11.84EE57 pKa = 4.25KK58 pKa = 10.74IRR60 pKa = 11.84FLDD63 pKa = 3.3SGGQYY68 pKa = 10.82GHH70 pKa = 6.87GPDD73 pKa = 3.84DD74 pKa = 3.89AKK76 pKa = 11.51VFIKK80 pKa = 10.54FEE82 pKa = 3.94ATKK85 pKa = 10.1QSKK88 pKa = 8.63VQEE91 pKa = 3.8QRR93 pKa = 11.84YY94 pKa = 8.96RR95 pKa = 11.84LIWSLSVEE103 pKa = 4.1DD104 pKa = 4.32QIVDD108 pKa = 3.46RR109 pKa = 11.84LLFGHH114 pKa = 7.48AFRR117 pKa = 11.84AEE119 pKa = 4.12VNHH122 pKa = 5.7FTEE125 pKa = 4.99TTSKK129 pKa = 9.58TGWSPLPGGYY139 pKa = 9.8SYY141 pKa = 11.42FNSTFNGHH149 pKa = 5.76VLATDD154 pKa = 3.87ASAFDD159 pKa = 3.51WTVQNWMVAMLYY171 pKa = 8.2DD172 pKa = 3.64TLMDD176 pKa = 3.53RR177 pKa = 11.84TRR179 pKa = 11.84GNTEE183 pKa = 3.45SYY185 pKa = 11.12KK186 pKa = 10.78RR187 pKa = 11.84MAFNRR192 pKa = 11.84IQQVLGPEE200 pKa = 4.29CVVRR204 pKa = 11.84LPNGRR209 pKa = 11.84RR210 pKa = 11.84FRR212 pKa = 11.84QRR214 pKa = 11.84VFGVMKK220 pKa = 10.56SGWLLTLMANTNVMLASSLCAWARR244 pKa = 11.84AAPAYY249 pKa = 10.5FEE251 pKa = 4.24ATPDD255 pKa = 3.35GLYY258 pKa = 10.32PDD260 pKa = 5.11MPLSWGMGDD269 pKa = 4.07DD270 pKa = 6.21VIMKK274 pKa = 9.04WDD276 pKa = 3.64PTLSPEE282 pKa = 4.15PYY284 pKa = 9.59VRR286 pKa = 11.84EE287 pKa = 4.05LNRR290 pKa = 11.84TGLKK294 pKa = 9.45VKK296 pKa = 10.2QYY298 pKa = 8.01SHH300 pKa = 6.02EE301 pKa = 4.15RR302 pKa = 11.84EE303 pKa = 3.8FAGFRR308 pKa = 11.84IQPSSGDD315 pKa = 3.49QQPTVTPIYY324 pKa = 8.24TAKK327 pKa = 10.12HH328 pKa = 4.34MTMIAYY334 pKa = 7.92TPRR337 pKa = 11.84EE338 pKa = 4.16RR339 pKa = 11.84LEE341 pKa = 4.16EE342 pKa = 3.76MAIAYY347 pKa = 8.84GLIYY351 pKa = 10.47ALSDD355 pKa = 3.49SASPQMKK362 pKa = 10.11KK363 pKa = 10.7FIDD366 pKa = 4.38DD367 pKa = 3.59YY368 pKa = 11.45SPYY371 pKa = 8.9GSSYY375 pKa = 9.79FEE377 pKa = 3.8HH378 pKa = 7.29WARR381 pKa = 11.84GQPLPGNDD389 pKa = 3.59GPRR392 pKa = 11.84VRR394 pKa = 11.84MQLDD398 pKa = 3.07NAYY401 pKa = 10.51ALARR405 pKa = 11.84TQDD408 pKa = 3.21QQ409 pKa = 3.26
MM1 pKa = 7.78DD2 pKa = 4.59FDD4 pKa = 4.08VRR6 pKa = 11.84VGRR9 pKa = 11.84ALKK12 pKa = 9.9EE13 pKa = 4.07CQPDD17 pKa = 3.7SVTGLGYY24 pKa = 10.23FRR26 pKa = 11.84NYY28 pKa = 7.2QTIGDD33 pKa = 4.21AFGHH37 pKa = 5.77EE38 pKa = 4.45CGFYY42 pKa = 10.61TNAHH46 pKa = 6.42NIAILVNIVRR56 pKa = 11.84EE57 pKa = 4.25KK58 pKa = 10.74IRR60 pKa = 11.84FLDD63 pKa = 3.3SGGQYY68 pKa = 10.82GHH70 pKa = 6.87GPDD73 pKa = 3.84DD74 pKa = 3.89AKK76 pKa = 11.51VFIKK80 pKa = 10.54FEE82 pKa = 3.94ATKK85 pKa = 10.1QSKK88 pKa = 8.63VQEE91 pKa = 3.8QRR93 pKa = 11.84YY94 pKa = 8.96RR95 pKa = 11.84LIWSLSVEE103 pKa = 4.1DD104 pKa = 4.32QIVDD108 pKa = 3.46RR109 pKa = 11.84LLFGHH114 pKa = 7.48AFRR117 pKa = 11.84AEE119 pKa = 4.12VNHH122 pKa = 5.7FTEE125 pKa = 4.99TTSKK129 pKa = 9.58TGWSPLPGGYY139 pKa = 9.8SYY141 pKa = 11.42FNSTFNGHH149 pKa = 5.76VLATDD154 pKa = 3.87ASAFDD159 pKa = 3.51WTVQNWMVAMLYY171 pKa = 8.2DD172 pKa = 3.64TLMDD176 pKa = 3.53RR177 pKa = 11.84TRR179 pKa = 11.84GNTEE183 pKa = 3.45SYY185 pKa = 11.12KK186 pKa = 10.78RR187 pKa = 11.84MAFNRR192 pKa = 11.84IQQVLGPEE200 pKa = 4.29CVVRR204 pKa = 11.84LPNGRR209 pKa = 11.84RR210 pKa = 11.84FRR212 pKa = 11.84QRR214 pKa = 11.84VFGVMKK220 pKa = 10.56SGWLLTLMANTNVMLASSLCAWARR244 pKa = 11.84AAPAYY249 pKa = 10.5FEE251 pKa = 4.24ATPDD255 pKa = 3.35GLYY258 pKa = 10.32PDD260 pKa = 5.11MPLSWGMGDD269 pKa = 4.07DD270 pKa = 6.21VIMKK274 pKa = 9.04WDD276 pKa = 3.64PTLSPEE282 pKa = 4.15PYY284 pKa = 9.59VRR286 pKa = 11.84EE287 pKa = 4.05LNRR290 pKa = 11.84TGLKK294 pKa = 9.45VKK296 pKa = 10.2QYY298 pKa = 8.01SHH300 pKa = 6.02EE301 pKa = 4.15RR302 pKa = 11.84EE303 pKa = 3.8FAGFRR308 pKa = 11.84IQPSSGDD315 pKa = 3.49QQPTVTPIYY324 pKa = 8.24TAKK327 pKa = 10.12HH328 pKa = 4.34MTMIAYY334 pKa = 7.92TPRR337 pKa = 11.84EE338 pKa = 4.16RR339 pKa = 11.84LEE341 pKa = 4.16EE342 pKa = 3.76MAIAYY347 pKa = 8.84GLIYY351 pKa = 10.47ALSDD355 pKa = 3.49SASPQMKK362 pKa = 10.11KK363 pKa = 10.7FIDD366 pKa = 4.38DD367 pKa = 3.59YY368 pKa = 11.45SPYY371 pKa = 8.9GSSYY375 pKa = 9.79FEE377 pKa = 3.8HH378 pKa = 7.29WARR381 pKa = 11.84GQPLPGNDD389 pKa = 3.59GPRR392 pKa = 11.84VRR394 pKa = 11.84MQLDD398 pKa = 3.07NAYY401 pKa = 10.51ALARR405 pKa = 11.84TQDD408 pKa = 3.21QQ409 pKa = 3.26
Molecular weight: 46.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEV8|A0A1L3KEV8_9VIRU Capsid protein OS=Wuhan heteroptera virus 2 OX=1923702 PE=3 SV=1
MM1 pKa = 6.87VRR3 pKa = 11.84SRR5 pKa = 11.84KK6 pKa = 7.89KK7 pKa = 8.98TKK9 pKa = 8.7PTSKK13 pKa = 10.46LVAEE17 pKa = 4.63VAAATKK23 pKa = 10.21AVQALKK29 pKa = 10.43IGTPARR35 pKa = 11.84KK36 pKa = 9.1RR37 pKa = 11.84KK38 pKa = 8.67PRR40 pKa = 11.84SRR42 pKa = 11.84VPRR45 pKa = 11.84QRR47 pKa = 11.84GAGNVYY53 pKa = 10.34SQANDD58 pKa = 3.02ATVVFTRR65 pKa = 11.84SEE67 pKa = 3.95MLEE70 pKa = 4.44AITVSDD76 pKa = 2.97QGTYY80 pKa = 10.36LKK82 pKa = 10.51HH83 pKa = 6.59FKK85 pKa = 8.7ITPATFPLLSKK96 pKa = 10.28IAQHH100 pKa = 6.62FDD102 pKa = 2.89QSQWLAVEE110 pKa = 4.81VYY112 pKa = 6.51WTSDD116 pKa = 2.75VGTTTGGSITYY127 pKa = 10.17GFDD130 pKa = 3.25WDD132 pKa = 3.73PTTAEE137 pKa = 4.55SKK139 pKa = 10.52NKK141 pKa = 8.31TRR143 pKa = 11.84AAITALTPSHH153 pKa = 6.41TGAVYY158 pKa = 10.26RR159 pKa = 11.84QTDD162 pKa = 3.88TAPLKK167 pKa = 10.72APKK170 pKa = 9.95NQLDD174 pKa = 3.56TLKK177 pKa = 10.07WYY179 pKa = 10.05KK180 pKa = 10.94HH181 pKa = 5.54NDD183 pKa = 2.92ATASADD189 pKa = 3.64SNLIAPCTVSVMASSTPKK207 pKa = 10.39LAIGVLWCRR216 pKa = 11.84YY217 pKa = 9.09RR218 pKa = 11.84IRR220 pKa = 11.84LRR222 pKa = 11.84GTTSXRR228 pKa = 11.84QLYY231 pKa = 9.34QLQTLSSVWTHH242 pKa = 7.07RR243 pKa = 11.84DD244 pKa = 3.3TQSAAWNTRR253 pKa = 11.84VWEE256 pKa = 4.12TDD258 pKa = 2.68EE259 pKa = 4.75LAPSKK264 pKa = 10.5PRR266 pKa = 11.84VYY268 pKa = 10.29AYY270 pKa = 9.75KK271 pKa = 10.37AATRR275 pKa = 11.84PVEE278 pKa = 3.9LPVRR282 pKa = 11.84SEE284 pKa = 3.62PTKK287 pKa = 10.56RR288 pKa = 11.84DD289 pKa = 3.26LSVQEE294 pKa = 4.72VKK296 pKa = 10.85QKK298 pKa = 11.67DD299 pKa = 3.53NMDD302 pKa = 3.96NVWSFNAGRR311 pKa = 11.84LPTPHH316 pKa = 6.89VIWTCQLYY324 pKa = 10.26VPVSFQPLAGTIADD338 pKa = 3.82ICFDD342 pKa = 3.47MQLVHH347 pKa = 6.72SHH349 pKa = 6.96DD350 pKa = 4.09RR351 pKa = 11.84LPTDD355 pKa = 3.15MTITLAPQALLEE367 pKa = 4.26SGYY370 pKa = 9.4VQEE373 pKa = 5.14STPSMYY379 pKa = 10.85LGGRR383 pKa = 11.84AMTVFSMLIGKK394 pKa = 9.62LMDD397 pKa = 3.21WTKK400 pKa = 11.16EE401 pKa = 3.7MSVLIGFEE409 pKa = 4.02FDD411 pKa = 4.53FNAFDD416 pKa = 3.92SSATQSLTFSYY427 pKa = 10.91NVTVRR432 pKa = 11.84LTSDD436 pKa = 3.7GASKK440 pKa = 10.91ASDD443 pKa = 3.45QQQLICKK450 pKa = 8.6SCQGVAEE457 pKa = 4.46IVPSRR462 pKa = 11.84PSSPGIPFYY471 pKa = 11.28GGVDD475 pKa = 3.11PGYY478 pKa = 10.11ISNGDD483 pKa = 3.7FVFVLPDD490 pKa = 3.56GSEE493 pKa = 4.07LPSRR497 pKa = 11.84PPINN501 pKa = 3.28
MM1 pKa = 6.87VRR3 pKa = 11.84SRR5 pKa = 11.84KK6 pKa = 7.89KK7 pKa = 8.98TKK9 pKa = 8.7PTSKK13 pKa = 10.46LVAEE17 pKa = 4.63VAAATKK23 pKa = 10.21AVQALKK29 pKa = 10.43IGTPARR35 pKa = 11.84KK36 pKa = 9.1RR37 pKa = 11.84KK38 pKa = 8.67PRR40 pKa = 11.84SRR42 pKa = 11.84VPRR45 pKa = 11.84QRR47 pKa = 11.84GAGNVYY53 pKa = 10.34SQANDD58 pKa = 3.02ATVVFTRR65 pKa = 11.84SEE67 pKa = 3.95MLEE70 pKa = 4.44AITVSDD76 pKa = 2.97QGTYY80 pKa = 10.36LKK82 pKa = 10.51HH83 pKa = 6.59FKK85 pKa = 8.7ITPATFPLLSKK96 pKa = 10.28IAQHH100 pKa = 6.62FDD102 pKa = 2.89QSQWLAVEE110 pKa = 4.81VYY112 pKa = 6.51WTSDD116 pKa = 2.75VGTTTGGSITYY127 pKa = 10.17GFDD130 pKa = 3.25WDD132 pKa = 3.73PTTAEE137 pKa = 4.55SKK139 pKa = 10.52NKK141 pKa = 8.31TRR143 pKa = 11.84AAITALTPSHH153 pKa = 6.41TGAVYY158 pKa = 10.26RR159 pKa = 11.84QTDD162 pKa = 3.88TAPLKK167 pKa = 10.72APKK170 pKa = 9.95NQLDD174 pKa = 3.56TLKK177 pKa = 10.07WYY179 pKa = 10.05KK180 pKa = 10.94HH181 pKa = 5.54NDD183 pKa = 2.92ATASADD189 pKa = 3.64SNLIAPCTVSVMASSTPKK207 pKa = 10.39LAIGVLWCRR216 pKa = 11.84YY217 pKa = 9.09RR218 pKa = 11.84IRR220 pKa = 11.84LRR222 pKa = 11.84GTTSXRR228 pKa = 11.84QLYY231 pKa = 9.34QLQTLSSVWTHH242 pKa = 7.07RR243 pKa = 11.84DD244 pKa = 3.3TQSAAWNTRR253 pKa = 11.84VWEE256 pKa = 4.12TDD258 pKa = 2.68EE259 pKa = 4.75LAPSKK264 pKa = 10.5PRR266 pKa = 11.84VYY268 pKa = 10.29AYY270 pKa = 9.75KK271 pKa = 10.37AATRR275 pKa = 11.84PVEE278 pKa = 3.9LPVRR282 pKa = 11.84SEE284 pKa = 3.62PTKK287 pKa = 10.56RR288 pKa = 11.84DD289 pKa = 3.26LSVQEE294 pKa = 4.72VKK296 pKa = 10.85QKK298 pKa = 11.67DD299 pKa = 3.53NMDD302 pKa = 3.96NVWSFNAGRR311 pKa = 11.84LPTPHH316 pKa = 6.89VIWTCQLYY324 pKa = 10.26VPVSFQPLAGTIADD338 pKa = 3.82ICFDD342 pKa = 3.47MQLVHH347 pKa = 6.72SHH349 pKa = 6.96DD350 pKa = 4.09RR351 pKa = 11.84LPTDD355 pKa = 3.15MTITLAPQALLEE367 pKa = 4.26SGYY370 pKa = 9.4VQEE373 pKa = 5.14STPSMYY379 pKa = 10.85LGGRR383 pKa = 11.84AMTVFSMLIGKK394 pKa = 9.62LMDD397 pKa = 3.21WTKK400 pKa = 11.16EE401 pKa = 3.7MSVLIGFEE409 pKa = 4.02FDD411 pKa = 4.53FNAFDD416 pKa = 3.92SSATQSLTFSYY427 pKa = 10.91NVTVRR432 pKa = 11.84LTSDD436 pKa = 3.7GASKK440 pKa = 10.91ASDD443 pKa = 3.45QQQLICKK450 pKa = 8.6SCQGVAEE457 pKa = 4.46IVPSRR462 pKa = 11.84PSSPGIPFYY471 pKa = 11.28GGVDD475 pKa = 3.11PGYY478 pKa = 10.11ISNGDD483 pKa = 3.7FVFVLPDD490 pKa = 3.56GSEE493 pKa = 4.07LPSRR497 pKa = 11.84PPINN501 pKa = 3.28
Molecular weight: 55.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1658 |
409 |
748 |
552.7 |
61.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.263 ± 0.208 | 1.206 ± 0.093 |
4.885 ± 0.584 | 4.403 ± 0.416 |
3.317 ± 0.741 | 5.911 ± 0.629 |
1.93 ± 0.131 | 4.282 ± 0.211 |
4.584 ± 0.381 | 8.323 ± 0.553 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.714 ± 0.451 | 3.739 ± 0.326 |
6.092 ± 0.28 | 5.609 ± 0.425 |
6.212 ± 0.346 | 9.59 ± 1.458 |
7.6 ± 0.808 | 5.971 ± 0.575 |
1.689 ± 0.322 | 3.619 ± 0.574 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
