
Methanococcus maripaludis (strain S2 / LL)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Methanomada group; Methanococci; Methanococcales; Methanococcaceae; Methanococcus; Methanococcus maripaludis
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
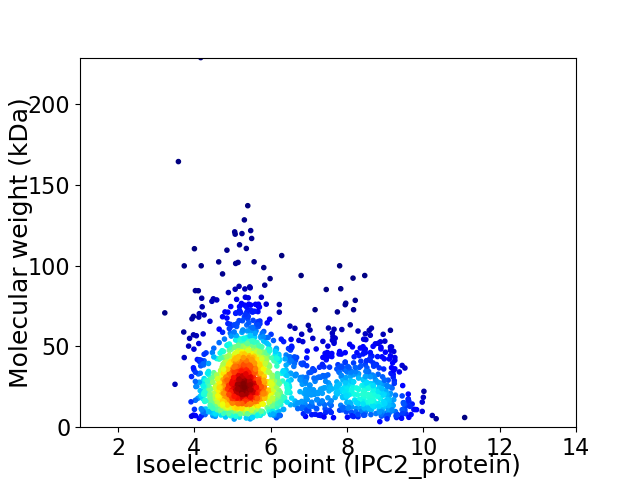
Virtual 2D-PAGE plot for 1722 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q6LX80|RFRNP_METMP RNA-free ribonuclease P OS=Methanococcus maripaludis (strain S2 / LL) OX=267377 GN=MMP1472 PE=3 SV=1
MM1 pKa = 7.53SAPSKK6 pKa = 10.75LIALFILTVLCTLTISYY23 pKa = 10.52GMVVDD28 pKa = 5.27DD29 pKa = 4.74PLIVVNSEE37 pKa = 4.26KK38 pKa = 10.35VDD40 pKa = 3.28KK41 pKa = 10.54TYY43 pKa = 11.55AEE45 pKa = 4.41LLMDD49 pKa = 4.04KK50 pKa = 10.41YY51 pKa = 9.11YY52 pKa = 9.89TKK54 pKa = 10.77RR55 pKa = 11.84SIEE58 pKa = 4.11VNSDD62 pKa = 2.45NDD64 pKa = 3.35IVVNEE69 pKa = 4.1NIIYY73 pKa = 9.39NVPALDD79 pKa = 3.51EE80 pKa = 4.67FEE82 pKa = 5.24INDD85 pKa = 3.91GKK87 pKa = 11.41GKK89 pKa = 10.41LLVEE93 pKa = 4.61FTKK96 pKa = 10.74SGEE99 pKa = 4.11SVTYY103 pKa = 10.82KK104 pKa = 10.06NFEE107 pKa = 4.04YY108 pKa = 10.78EE109 pKa = 4.27EE110 pKa = 4.63EE111 pKa = 4.22IEE113 pKa = 5.0SDD115 pKa = 3.89DD116 pKa = 4.38EE117 pKa = 5.28GDD119 pKa = 3.41EE120 pKa = 4.14VTFMGKK126 pKa = 7.51TYY128 pKa = 11.21KK129 pKa = 10.59LIEE132 pKa = 4.18YY133 pKa = 10.53DD134 pKa = 3.63EE135 pKa = 4.88DD136 pKa = 3.69EE137 pKa = 5.89KK138 pKa = 11.14IVLGNKK144 pKa = 9.37VEE146 pKa = 5.62DD147 pKa = 3.53IDD149 pKa = 3.94TTEE152 pKa = 3.89EE153 pKa = 3.73FSYY156 pKa = 10.85DD157 pKa = 2.7IYY159 pKa = 10.54TFKK162 pKa = 10.77IIGKK166 pKa = 9.36DD167 pKa = 3.17STGEE171 pKa = 3.99VLVSVYY177 pKa = 10.68EE178 pKa = 4.12GTDD181 pKa = 2.95RR182 pKa = 11.84IEE184 pKa = 4.1NVKK187 pKa = 9.8MYY189 pKa = 10.36PDD191 pKa = 3.25DD192 pKa = 3.74TYY194 pKa = 10.98FVSGSTVSIYY204 pKa = 10.45YY205 pKa = 10.13DD206 pKa = 3.53EE207 pKa = 4.52YY208 pKa = 11.59LEE210 pKa = 5.08SGDD213 pKa = 3.15TAYY216 pKa = 10.74FFFEE220 pKa = 4.71IYY222 pKa = 10.43DD223 pKa = 3.64SLEE226 pKa = 4.21FEE228 pKa = 4.88DD229 pKa = 4.88GKK231 pKa = 11.28SISGYY236 pKa = 10.55SDD238 pKa = 3.82FDD240 pKa = 4.03TEE242 pKa = 5.35IDD244 pKa = 3.68GQTITLEE251 pKa = 3.9YY252 pKa = 10.62DD253 pKa = 3.33NPKK256 pKa = 10.4SLSEE260 pKa = 4.12NFGLLNYY267 pKa = 9.63KK268 pKa = 10.21VNLVDD273 pKa = 5.12VNGEE277 pKa = 3.9DD278 pKa = 3.15LTAFFTVTQNLNYY291 pKa = 9.96EE292 pKa = 4.07YY293 pKa = 11.41DD294 pKa = 3.44MDD296 pKa = 4.37EE297 pKa = 3.88GTKK300 pKa = 10.72YY301 pKa = 10.65FGNNIFAVKK310 pKa = 10.43LEE312 pKa = 4.08NDD314 pKa = 4.53EE315 pKa = 4.21EE316 pKa = 5.05DD317 pKa = 3.71DD318 pKa = 4.12WDD320 pKa = 5.14DD321 pKa = 4.41EE322 pKa = 4.87LYY324 pKa = 10.68LYY326 pKa = 10.81KK327 pKa = 10.82NNKK330 pKa = 9.62KK331 pKa = 9.55YY332 pKa = 11.17DD333 pKa = 3.93KK334 pKa = 10.71IVDD337 pKa = 3.86YY338 pKa = 10.78QGSVQLVDD346 pKa = 3.41QDD348 pKa = 4.23EE349 pKa = 4.48LLSASSDD356 pKa = 3.84LILIGGPVSNKK367 pKa = 6.09VTEE370 pKa = 4.66SIQNSLNIEE379 pKa = 4.06VTNDD383 pKa = 3.13YY384 pKa = 10.4PGEE387 pKa = 4.26GKK389 pKa = 10.73GVIQTITNPNNAEE402 pKa = 3.98STILVLAGSDD412 pKa = 3.51RR413 pKa = 11.84EE414 pKa = 4.43GTKK417 pKa = 10.61ACVLALNQGLYY428 pKa = 10.46SGTGALTVKK437 pKa = 9.67LTGEE441 pKa = 4.39DD442 pKa = 3.56SVSVII447 pKa = 4.39
MM1 pKa = 7.53SAPSKK6 pKa = 10.75LIALFILTVLCTLTISYY23 pKa = 10.52GMVVDD28 pKa = 5.27DD29 pKa = 4.74PLIVVNSEE37 pKa = 4.26KK38 pKa = 10.35VDD40 pKa = 3.28KK41 pKa = 10.54TYY43 pKa = 11.55AEE45 pKa = 4.41LLMDD49 pKa = 4.04KK50 pKa = 10.41YY51 pKa = 9.11YY52 pKa = 9.89TKK54 pKa = 10.77RR55 pKa = 11.84SIEE58 pKa = 4.11VNSDD62 pKa = 2.45NDD64 pKa = 3.35IVVNEE69 pKa = 4.1NIIYY73 pKa = 9.39NVPALDD79 pKa = 3.51EE80 pKa = 4.67FEE82 pKa = 5.24INDD85 pKa = 3.91GKK87 pKa = 11.41GKK89 pKa = 10.41LLVEE93 pKa = 4.61FTKK96 pKa = 10.74SGEE99 pKa = 4.11SVTYY103 pKa = 10.82KK104 pKa = 10.06NFEE107 pKa = 4.04YY108 pKa = 10.78EE109 pKa = 4.27EE110 pKa = 4.63EE111 pKa = 4.22IEE113 pKa = 5.0SDD115 pKa = 3.89DD116 pKa = 4.38EE117 pKa = 5.28GDD119 pKa = 3.41EE120 pKa = 4.14VTFMGKK126 pKa = 7.51TYY128 pKa = 11.21KK129 pKa = 10.59LIEE132 pKa = 4.18YY133 pKa = 10.53DD134 pKa = 3.63EE135 pKa = 4.88DD136 pKa = 3.69EE137 pKa = 5.89KK138 pKa = 11.14IVLGNKK144 pKa = 9.37VEE146 pKa = 5.62DD147 pKa = 3.53IDD149 pKa = 3.94TTEE152 pKa = 3.89EE153 pKa = 3.73FSYY156 pKa = 10.85DD157 pKa = 2.7IYY159 pKa = 10.54TFKK162 pKa = 10.77IIGKK166 pKa = 9.36DD167 pKa = 3.17STGEE171 pKa = 3.99VLVSVYY177 pKa = 10.68EE178 pKa = 4.12GTDD181 pKa = 2.95RR182 pKa = 11.84IEE184 pKa = 4.1NVKK187 pKa = 9.8MYY189 pKa = 10.36PDD191 pKa = 3.25DD192 pKa = 3.74TYY194 pKa = 10.98FVSGSTVSIYY204 pKa = 10.45YY205 pKa = 10.13DD206 pKa = 3.53EE207 pKa = 4.52YY208 pKa = 11.59LEE210 pKa = 5.08SGDD213 pKa = 3.15TAYY216 pKa = 10.74FFFEE220 pKa = 4.71IYY222 pKa = 10.43DD223 pKa = 3.64SLEE226 pKa = 4.21FEE228 pKa = 4.88DD229 pKa = 4.88GKK231 pKa = 11.28SISGYY236 pKa = 10.55SDD238 pKa = 3.82FDD240 pKa = 4.03TEE242 pKa = 5.35IDD244 pKa = 3.68GQTITLEE251 pKa = 3.9YY252 pKa = 10.62DD253 pKa = 3.33NPKK256 pKa = 10.4SLSEE260 pKa = 4.12NFGLLNYY267 pKa = 9.63KK268 pKa = 10.21VNLVDD273 pKa = 5.12VNGEE277 pKa = 3.9DD278 pKa = 3.15LTAFFTVTQNLNYY291 pKa = 9.96EE292 pKa = 4.07YY293 pKa = 11.41DD294 pKa = 3.44MDD296 pKa = 4.37EE297 pKa = 3.88GTKK300 pKa = 10.72YY301 pKa = 10.65FGNNIFAVKK310 pKa = 10.43LEE312 pKa = 4.08NDD314 pKa = 4.53EE315 pKa = 4.21EE316 pKa = 5.05DD317 pKa = 3.71DD318 pKa = 4.12WDD320 pKa = 5.14DD321 pKa = 4.41EE322 pKa = 4.87LYY324 pKa = 10.68LYY326 pKa = 10.81KK327 pKa = 10.82NNKK330 pKa = 9.62KK331 pKa = 9.55YY332 pKa = 11.17DD333 pKa = 3.93KK334 pKa = 10.71IVDD337 pKa = 3.86YY338 pKa = 10.78QGSVQLVDD346 pKa = 3.41QDD348 pKa = 4.23EE349 pKa = 4.48LLSASSDD356 pKa = 3.84LILIGGPVSNKK367 pKa = 6.09VTEE370 pKa = 4.66SIQNSLNIEE379 pKa = 4.06VTNDD383 pKa = 3.13YY384 pKa = 10.4PGEE387 pKa = 4.26GKK389 pKa = 10.73GVIQTITNPNNAEE402 pKa = 3.98STILVLAGSDD412 pKa = 3.51RR413 pKa = 11.84EE414 pKa = 4.43GTKK417 pKa = 10.61ACVLALNQGLYY428 pKa = 10.46SGTGALTVKK437 pKa = 9.67LTGEE441 pKa = 4.39DD442 pKa = 3.56SVSVII447 pKa = 4.39
Molecular weight: 50.23 kDa
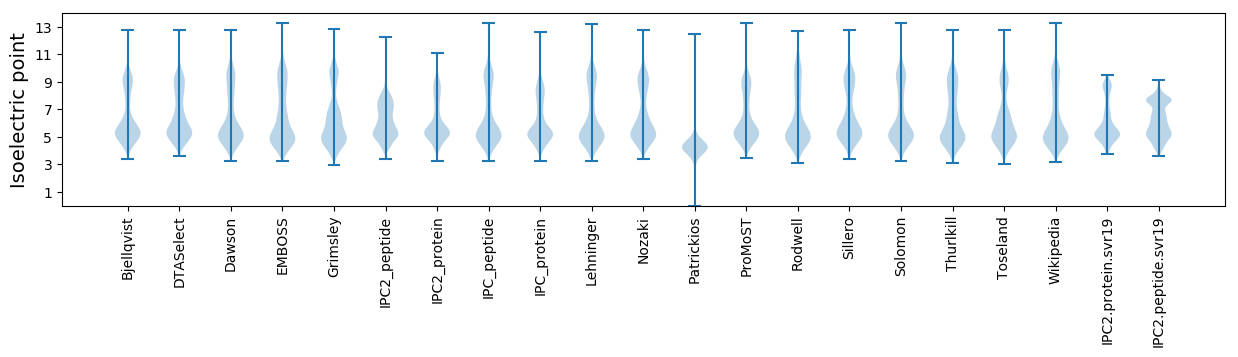
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6M0Y3|Q6M0Y3_METMP Threonine synthase OS=Methanococcus maripaludis (strain S2 / LL) OX=267377 GN=thrC PE=3 SV=1
MM1 pKa = 7.77AGNKK5 pKa = 9.37PLGKK9 pKa = 10.1KK10 pKa = 9.19IRR12 pKa = 11.84LAKK15 pKa = 10.25ALKK18 pKa = 9.04QNRR21 pKa = 11.84RR22 pKa = 11.84VPMFAIARR30 pKa = 11.84TKK32 pKa = 10.89GSVKK36 pKa = 8.79QHH38 pKa = 5.52PKK40 pKa = 7.54MRR42 pKa = 11.84HH43 pKa = 3.6WRR45 pKa = 11.84RR46 pKa = 11.84KK47 pKa = 7.42NLKK50 pKa = 9.64KK51 pKa = 10.79
MM1 pKa = 7.77AGNKK5 pKa = 9.37PLGKK9 pKa = 10.1KK10 pKa = 9.19IRR12 pKa = 11.84LAKK15 pKa = 10.25ALKK18 pKa = 9.04QNRR21 pKa = 11.84RR22 pKa = 11.84VPMFAIARR30 pKa = 11.84TKK32 pKa = 10.89GSVKK36 pKa = 8.79QHH38 pKa = 5.52PKK40 pKa = 7.54MRR42 pKa = 11.84HH43 pKa = 3.6WRR45 pKa = 11.84RR46 pKa = 11.84KK47 pKa = 7.42NLKK50 pKa = 9.64KK51 pKa = 10.79
Molecular weight: 6.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
490599 |
32 |
2076 |
284.9 |
31.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.809 ± 0.071 | 1.367 ± 0.04 |
5.602 ± 0.048 | 8.131 ± 0.06 |
4.2 ± 0.05 | 6.753 ± 0.049 |
1.501 ± 0.023 | 9.401 ± 0.059 |
8.849 ± 0.067 | 9.088 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.623 ± 0.031 | 5.637 ± 0.064 |
3.398 ± 0.031 | 1.871 ± 0.024 |
3.096 ± 0.043 | 6.257 ± 0.055 |
4.965 ± 0.044 | 6.982 ± 0.052 |
0.65 ± 0.019 | 3.82 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
