
Bovine alphaherpesvirus 1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Varicellovirus
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
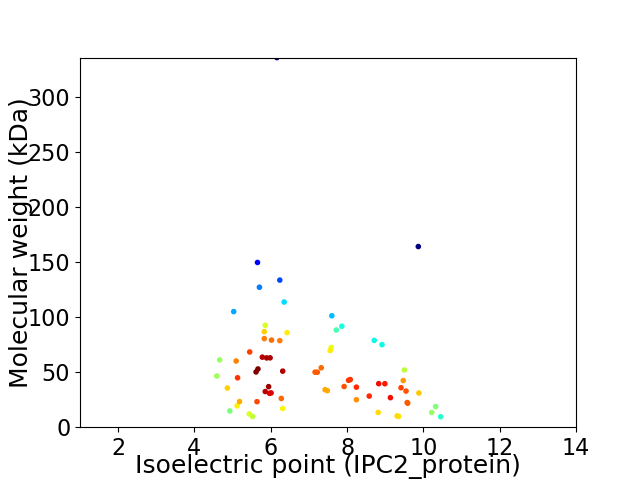
Virtual 2D-PAGE plot for 71 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q76PK0|Q76PK0_9ALPH Capsid scaffolding protein OS=Bovine alphaherpesvirus 1 OX=10320 GN=UL26 PE=3 SV=1
MM1 pKa = 7.36PAARR5 pKa = 11.84TGTLAAVALILLCGAAVLGRR25 pKa = 11.84PAPDD29 pKa = 4.02DD30 pKa = 3.8LCFADD35 pKa = 4.08VRR37 pKa = 11.84RR38 pKa = 11.84TGMAPSRR45 pKa = 11.84PLGPVLNLAASDD57 pKa = 3.54LTSRR61 pKa = 11.84VSVRR65 pKa = 11.84AVDD68 pKa = 3.92ASRR71 pKa = 11.84GCALALLDD79 pKa = 3.67MAEE82 pKa = 4.3TVVPGGPRR90 pKa = 11.84AADD93 pKa = 3.39VVDD96 pKa = 4.23VGWAYY101 pKa = 11.0QDD103 pKa = 3.56GDD105 pKa = 4.21CMVPLAYY112 pKa = 9.55RR113 pKa = 11.84QYY115 pKa = 10.87FNCTGGALPGQNVCAGLSEE134 pKa = 4.01TRR136 pKa = 11.84IRR138 pKa = 11.84GGFGTSDD145 pKa = 3.4YY146 pKa = 11.42ALYY149 pKa = 8.63GTSLVLRR156 pKa = 11.84PGLYY160 pKa = 10.4DD161 pKa = 3.67RR162 pKa = 11.84GTYY165 pKa = 9.46IYY167 pKa = 10.27FLGYY171 pKa = 10.52GPDD174 pKa = 4.19DD175 pKa = 3.88IYY177 pKa = 11.6VGSVTLMVGADD188 pKa = 3.09IHH190 pKa = 6.87KK191 pKa = 9.82YY192 pKa = 9.69PCGLDD197 pKa = 2.98RR198 pKa = 11.84GLGVALHH205 pKa = 6.47HH206 pKa = 6.78KK207 pKa = 10.03SGPARR212 pKa = 11.84PLTEE216 pKa = 5.8DD217 pKa = 4.34DD218 pKa = 4.19ATGDD222 pKa = 3.7WACGCFPALVEE233 pKa = 4.6VDD235 pKa = 4.14AVWGNVSAAEE245 pKa = 4.6LGLADD250 pKa = 5.37PIDD253 pKa = 4.03YY254 pKa = 10.57ADD256 pKa = 3.77EE257 pKa = 4.71GGEE260 pKa = 4.19VEE262 pKa = 4.59VLEE265 pKa = 5.2DD266 pKa = 3.65EE267 pKa = 5.03AGSASGNLPQDD278 pKa = 4.52DD279 pKa = 5.17PDD281 pKa = 5.43PDD283 pKa = 4.12LADD286 pKa = 3.86CRR288 pKa = 11.84TVGLFSEE295 pKa = 4.51SDD297 pKa = 3.42MFRR300 pKa = 11.84TASGPEE306 pKa = 3.85SLLIGAVAKK315 pKa = 10.63DD316 pKa = 3.9VLTVPLNLPPGRR328 pKa = 11.84SYY330 pKa = 11.24EE331 pKa = 4.08ALRR334 pKa = 11.84NASLEE339 pKa = 4.36CNSRR343 pKa = 11.84PRR345 pKa = 11.84EE346 pKa = 4.01TGDD349 pKa = 3.21AAVVVMSLQEE359 pKa = 3.81PARR362 pKa = 11.84LEE364 pKa = 3.95RR365 pKa = 11.84RR366 pKa = 11.84PDD368 pKa = 3.2ARR370 pKa = 11.84ATDD373 pKa = 3.88PEE375 pKa = 4.53FGLFGLPDD383 pKa = 4.19DD384 pKa = 4.18PAVRR388 pKa = 11.84RR389 pKa = 11.84GILIGLAIALLVLLFSLVIVLVCACRR415 pKa = 11.84LARR418 pKa = 11.84AAKK421 pKa = 8.82AARR424 pKa = 11.84RR425 pKa = 11.84ARR427 pKa = 11.84AATFAKK433 pKa = 10.28SNPAYY438 pKa = 10.29EE439 pKa = 4.49PMLSVV444 pKa = 3.74
MM1 pKa = 7.36PAARR5 pKa = 11.84TGTLAAVALILLCGAAVLGRR25 pKa = 11.84PAPDD29 pKa = 4.02DD30 pKa = 3.8LCFADD35 pKa = 4.08VRR37 pKa = 11.84RR38 pKa = 11.84TGMAPSRR45 pKa = 11.84PLGPVLNLAASDD57 pKa = 3.54LTSRR61 pKa = 11.84VSVRR65 pKa = 11.84AVDD68 pKa = 3.92ASRR71 pKa = 11.84GCALALLDD79 pKa = 3.67MAEE82 pKa = 4.3TVVPGGPRR90 pKa = 11.84AADD93 pKa = 3.39VVDD96 pKa = 4.23VGWAYY101 pKa = 11.0QDD103 pKa = 3.56GDD105 pKa = 4.21CMVPLAYY112 pKa = 9.55RR113 pKa = 11.84QYY115 pKa = 10.87FNCTGGALPGQNVCAGLSEE134 pKa = 4.01TRR136 pKa = 11.84IRR138 pKa = 11.84GGFGTSDD145 pKa = 3.4YY146 pKa = 11.42ALYY149 pKa = 8.63GTSLVLRR156 pKa = 11.84PGLYY160 pKa = 10.4DD161 pKa = 3.67RR162 pKa = 11.84GTYY165 pKa = 9.46IYY167 pKa = 10.27FLGYY171 pKa = 10.52GPDD174 pKa = 4.19DD175 pKa = 3.88IYY177 pKa = 11.6VGSVTLMVGADD188 pKa = 3.09IHH190 pKa = 6.87KK191 pKa = 9.82YY192 pKa = 9.69PCGLDD197 pKa = 2.98RR198 pKa = 11.84GLGVALHH205 pKa = 6.47HH206 pKa = 6.78KK207 pKa = 10.03SGPARR212 pKa = 11.84PLTEE216 pKa = 5.8DD217 pKa = 4.34DD218 pKa = 4.19ATGDD222 pKa = 3.7WACGCFPALVEE233 pKa = 4.6VDD235 pKa = 4.14AVWGNVSAAEE245 pKa = 4.6LGLADD250 pKa = 5.37PIDD253 pKa = 4.03YY254 pKa = 10.57ADD256 pKa = 3.77EE257 pKa = 4.71GGEE260 pKa = 4.19VEE262 pKa = 4.59VLEE265 pKa = 5.2DD266 pKa = 3.65EE267 pKa = 5.03AGSASGNLPQDD278 pKa = 4.52DD279 pKa = 5.17PDD281 pKa = 5.43PDD283 pKa = 4.12LADD286 pKa = 3.86CRR288 pKa = 11.84TVGLFSEE295 pKa = 4.51SDD297 pKa = 3.42MFRR300 pKa = 11.84TASGPEE306 pKa = 3.85SLLIGAVAKK315 pKa = 10.63DD316 pKa = 3.9VLTVPLNLPPGRR328 pKa = 11.84SYY330 pKa = 11.24EE331 pKa = 4.08ALRR334 pKa = 11.84NASLEE339 pKa = 4.36CNSRR343 pKa = 11.84PRR345 pKa = 11.84EE346 pKa = 4.01TGDD349 pKa = 3.21AAVVVMSLQEE359 pKa = 3.81PARR362 pKa = 11.84LEE364 pKa = 3.95RR365 pKa = 11.84RR366 pKa = 11.84PDD368 pKa = 3.2ARR370 pKa = 11.84ATDD373 pKa = 3.88PEE375 pKa = 4.53FGLFGLPDD383 pKa = 4.19DD384 pKa = 4.18PAVRR388 pKa = 11.84RR389 pKa = 11.84GILIGLAIALLVLLFSLVIVLVCACRR415 pKa = 11.84LARR418 pKa = 11.84AAKK421 pKa = 8.82AARR424 pKa = 11.84RR425 pKa = 11.84ARR427 pKa = 11.84AATFAKK433 pKa = 10.28SNPAYY438 pKa = 10.29EE439 pKa = 4.49PMLSVV444 pKa = 3.74
Molecular weight: 46.56 kDa
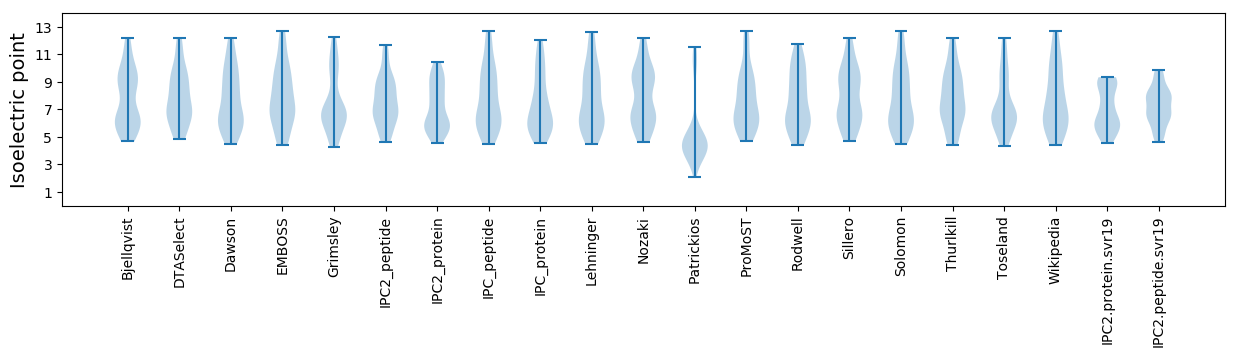
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q65546|Q65546_9ALPH Protein V57 OS=Bovine alphaherpesvirus 1 OX=10320 GN=UL3.5 PE=4 SV=1
MM1 pKa = 7.51RR2 pKa = 11.84RR3 pKa = 11.84VEE5 pKa = 4.23PVLSLLAAGGWQVSAAAAASASAPADD31 pKa = 3.84DD32 pKa = 5.17GEE34 pKa = 4.57RR35 pKa = 11.84SSGPALAGPGLAAAGLGPNPDD56 pKa = 3.31CVAFDD61 pKa = 3.83TLFMVSSIDD70 pKa = 3.47EE71 pKa = 4.18LGRR74 pKa = 11.84RR75 pKa = 11.84QLTDD79 pKa = 3.94TIRR82 pKa = 11.84KK83 pKa = 8.84DD84 pKa = 4.08LKK86 pKa = 10.15RR87 pKa = 11.84TLAKK91 pKa = 9.56YY92 pKa = 8.7TIACTKK98 pKa = 9.43TSSFSGAQARR108 pKa = 11.84GLSARR113 pKa = 11.84AARR116 pKa = 11.84AAARR120 pKa = 11.84GLRR123 pKa = 11.84SNKK126 pKa = 9.3SLQMFLLCQRR136 pKa = 11.84AHH138 pKa = 5.86AQRR141 pKa = 11.84VRR143 pKa = 11.84DD144 pKa = 3.71QLHH147 pKa = 6.1SVIQSRR153 pKa = 11.84KK154 pKa = 6.64PRR156 pKa = 11.84KK157 pKa = 8.98YY158 pKa = 8.55YY159 pKa = 9.93TRR161 pKa = 11.84SSDD164 pKa = 2.86GGTRR168 pKa = 11.84PAVPVYY174 pKa = 10.58VYY176 pKa = 9.53EE177 pKa = 4.82FSAAGPVYY185 pKa = 10.26IHH187 pKa = 7.5RR188 pKa = 11.84DD189 pKa = 3.24NTLGPDD195 pKa = 3.67SRR197 pKa = 11.84PGEE200 pKa = 4.06QPHH203 pKa = 6.23RR204 pKa = 4.12
MM1 pKa = 7.51RR2 pKa = 11.84RR3 pKa = 11.84VEE5 pKa = 4.23PVLSLLAAGGWQVSAAAAASASAPADD31 pKa = 3.84DD32 pKa = 5.17GEE34 pKa = 4.57RR35 pKa = 11.84SSGPALAGPGLAAAGLGPNPDD56 pKa = 3.31CVAFDD61 pKa = 3.83TLFMVSSIDD70 pKa = 3.47EE71 pKa = 4.18LGRR74 pKa = 11.84RR75 pKa = 11.84QLTDD79 pKa = 3.94TIRR82 pKa = 11.84KK83 pKa = 8.84DD84 pKa = 4.08LKK86 pKa = 10.15RR87 pKa = 11.84TLAKK91 pKa = 9.56YY92 pKa = 8.7TIACTKK98 pKa = 9.43TSSFSGAQARR108 pKa = 11.84GLSARR113 pKa = 11.84AARR116 pKa = 11.84AAARR120 pKa = 11.84GLRR123 pKa = 11.84SNKK126 pKa = 9.3SLQMFLLCQRR136 pKa = 11.84AHH138 pKa = 5.86AQRR141 pKa = 11.84VRR143 pKa = 11.84DD144 pKa = 3.71QLHH147 pKa = 6.1SVIQSRR153 pKa = 11.84KK154 pKa = 6.64PRR156 pKa = 11.84KK157 pKa = 8.98YY158 pKa = 8.55YY159 pKa = 9.93TRR161 pKa = 11.84SSDD164 pKa = 2.86GGTRR168 pKa = 11.84PAVPVYY174 pKa = 10.58VYY176 pKa = 9.53EE177 pKa = 4.82FSAAGPVYY185 pKa = 10.26IHH187 pKa = 7.5RR188 pKa = 11.84DD189 pKa = 3.24NTLGPDD195 pKa = 3.67SRR197 pKa = 11.84PGEE200 pKa = 4.06QPHH203 pKa = 6.23RR204 pKa = 4.12
Molecular weight: 21.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
38093 |
87 |
3291 |
536.5 |
56.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
19.153 ± 0.739 | 1.848 ± 0.133 |
5.046 ± 0.139 | 5.321 ± 0.173 |
3.137 ± 0.239 | 9.044 ± 0.367 |
1.869 ± 0.126 | 1.88 ± 0.112 |
1.328 ± 0.14 | 9.815 ± 0.289 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.467 ± 0.104 | 1.683 ± 0.159 |
7.807 ± 0.536 | 2.197 ± 0.103 |
9.521 ± 0.368 | 5.119 ± 0.181 |
3.906 ± 0.169 | 6.573 ± 0.261 |
0.948 ± 0.052 | 2.336 ± 0.188 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
