
Wigeon coronavirus HKU20
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Nidovirales; Cornidovirineae; Coronaviridae; Orthocoronavirinae; Deltacoronavirus; Andecovirus
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
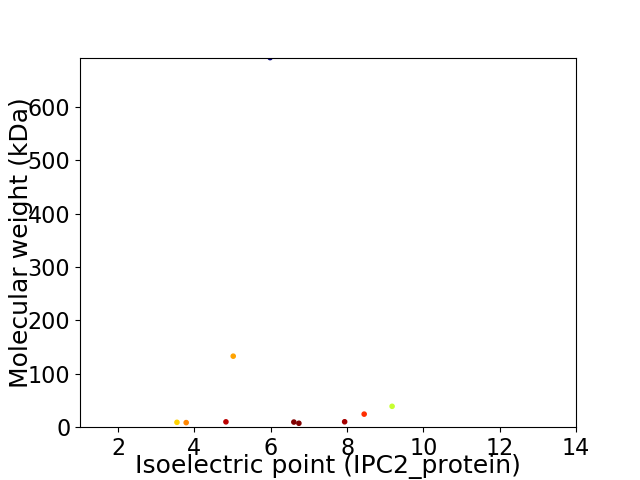
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H9BR31|H9BR31_9NIDO NS7b protein OS=Wigeon coronavirus HKU20 OX=1159908 GN=NS7b PE=4 SV=1
MM1 pKa = 7.35SLVIDD6 pKa = 4.5GDD8 pKa = 4.04PLQVADD14 pKa = 4.49VLRR17 pKa = 11.84QLSLIFHH24 pKa = 6.14TPIYY28 pKa = 10.43DD29 pKa = 3.62CNLRR33 pKa = 11.84LHH35 pKa = 5.78WVEE38 pKa = 5.34GVPCTLSFCEE48 pKa = 4.08LCYY51 pKa = 11.12DD52 pKa = 4.22FLQPIQADD60 pKa = 3.64SPGFSDD66 pKa = 5.2SSTSPDD72 pKa = 3.47FVDD75 pKa = 3.75SQQ77 pKa = 3.64
MM1 pKa = 7.35SLVIDD6 pKa = 4.5GDD8 pKa = 4.04PLQVADD14 pKa = 4.49VLRR17 pKa = 11.84QLSLIFHH24 pKa = 6.14TPIYY28 pKa = 10.43DD29 pKa = 3.62CNLRR33 pKa = 11.84LHH35 pKa = 5.78WVEE38 pKa = 5.34GVPCTLSFCEE48 pKa = 4.08LCYY51 pKa = 11.12DD52 pKa = 4.22FLQPIQADD60 pKa = 3.64SPGFSDD66 pKa = 5.2SSTSPDD72 pKa = 3.47FVDD75 pKa = 3.75SQQ77 pKa = 3.64
Molecular weight: 8.61 kDa
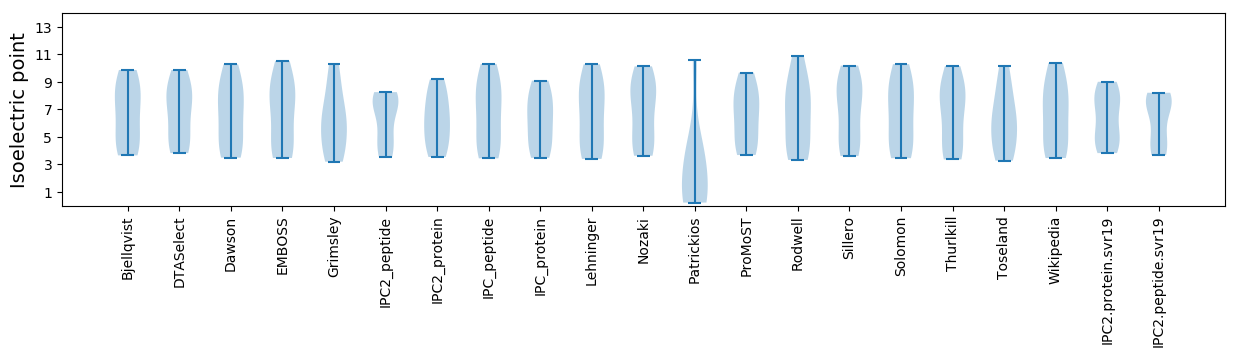
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H9BR30|H9BR30_9NIDO NS7a protein OS=Wigeon coronavirus HKU20 OX=1159908 GN=NS7a PE=4 SV=1
MM1 pKa = 7.65AVPSVPKK8 pKa = 10.31ADD10 pKa = 4.02CSWFQIIKK18 pKa = 9.74AQNKK22 pKa = 8.62KK23 pKa = 9.03VQPLAFKK30 pKa = 11.01GDD32 pKa = 3.7GVPINTTLPKK42 pKa = 10.19EE43 pKa = 4.09MCHH46 pKa = 6.72GYY48 pKa = 7.94WLRR51 pKa = 11.84YY52 pKa = 9.55KK53 pKa = 10.2RR54 pKa = 11.84VKK56 pKa = 10.32PGGAPLPPSYY66 pKa = 10.98SFYY69 pKa = 10.38YY70 pKa = 9.98TGTGPRR76 pKa = 11.84ATLKK80 pKa = 10.9YY81 pKa = 10.97GEE83 pKa = 4.99LQDD86 pKa = 5.8SDD88 pKa = 3.91NQDD91 pKa = 2.9NVNRR95 pKa = 11.84VLWVKK100 pKa = 10.85SPDD103 pKa = 3.43ADD105 pKa = 3.57TSIKK109 pKa = 10.42PKK111 pKa = 9.23VAKK114 pKa = 10.22RR115 pKa = 11.84NPDD118 pKa = 3.19KK119 pKa = 11.25HH120 pKa = 6.39PLLPLRR126 pKa = 11.84FKK128 pKa = 10.93PGDD131 pKa = 3.56GPVQGFRR138 pKa = 11.84IDD140 pKa = 3.65PFQSRR145 pKa = 11.84GRR147 pKa = 11.84SVEE150 pKa = 4.02RR151 pKa = 11.84GQQDD155 pKa = 2.74RR156 pKa = 11.84RR157 pKa = 11.84AQSAEE162 pKa = 3.67PQRR165 pKa = 11.84PKK167 pKa = 10.02QRR169 pKa = 11.84EE170 pKa = 3.83RR171 pKa = 11.84SRR173 pKa = 11.84SAPSRR178 pKa = 11.84AAPKK182 pKa = 9.68QHH184 pKa = 7.29AIPKK188 pKa = 9.73RR189 pKa = 11.84VLLKK193 pKa = 10.81GKK195 pKa = 8.28TISGTFGKK203 pKa = 10.44RR204 pKa = 11.84SPAKK208 pKa = 10.52ANVGSPDD215 pKa = 3.57TEE217 pKa = 4.07KK218 pKa = 10.41TGLGDD223 pKa = 3.84PRR225 pKa = 11.84LMALMRR231 pKa = 11.84FVPGTQEE238 pKa = 3.95LLFAGHH244 pKa = 7.68LKK246 pKa = 10.6HH247 pKa = 6.84KK248 pKa = 7.01MQPEE252 pKa = 4.91GVQLTFTYY260 pKa = 10.56DD261 pKa = 2.96ITVKK265 pKa = 10.4RR266 pKa = 11.84DD267 pKa = 3.12NKK269 pKa = 10.68DD270 pKa = 2.6FDD272 pKa = 5.24RR273 pKa = 11.84IVEE276 pKa = 4.13ALNSVVDD283 pKa = 3.63MVYY286 pKa = 10.43EE287 pKa = 3.89PEE289 pKa = 4.14GEE291 pKa = 3.95IRR293 pKa = 11.84KK294 pKa = 8.97DD295 pKa = 3.46QQSKK299 pKa = 11.15KK300 pKa = 9.59DD301 pKa = 3.58TQPKK305 pKa = 9.62KK306 pKa = 10.07KK307 pKa = 10.21AEE309 pKa = 4.3KK310 pKa = 8.91KK311 pKa = 9.19TPTNPQKK318 pKa = 10.9QPDD321 pKa = 4.11KK322 pKa = 10.48PVATIVDD329 pKa = 3.74NGDD332 pKa = 3.44SQVINWGTDD341 pKa = 3.19PAEE344 pKa = 4.29GLEE347 pKa = 4.34SFSS350 pKa = 4.03
MM1 pKa = 7.65AVPSVPKK8 pKa = 10.31ADD10 pKa = 4.02CSWFQIIKK18 pKa = 9.74AQNKK22 pKa = 8.62KK23 pKa = 9.03VQPLAFKK30 pKa = 11.01GDD32 pKa = 3.7GVPINTTLPKK42 pKa = 10.19EE43 pKa = 4.09MCHH46 pKa = 6.72GYY48 pKa = 7.94WLRR51 pKa = 11.84YY52 pKa = 9.55KK53 pKa = 10.2RR54 pKa = 11.84VKK56 pKa = 10.32PGGAPLPPSYY66 pKa = 10.98SFYY69 pKa = 10.38YY70 pKa = 9.98TGTGPRR76 pKa = 11.84ATLKK80 pKa = 10.9YY81 pKa = 10.97GEE83 pKa = 4.99LQDD86 pKa = 5.8SDD88 pKa = 3.91NQDD91 pKa = 2.9NVNRR95 pKa = 11.84VLWVKK100 pKa = 10.85SPDD103 pKa = 3.43ADD105 pKa = 3.57TSIKK109 pKa = 10.42PKK111 pKa = 9.23VAKK114 pKa = 10.22RR115 pKa = 11.84NPDD118 pKa = 3.19KK119 pKa = 11.25HH120 pKa = 6.39PLLPLRR126 pKa = 11.84FKK128 pKa = 10.93PGDD131 pKa = 3.56GPVQGFRR138 pKa = 11.84IDD140 pKa = 3.65PFQSRR145 pKa = 11.84GRR147 pKa = 11.84SVEE150 pKa = 4.02RR151 pKa = 11.84GQQDD155 pKa = 2.74RR156 pKa = 11.84RR157 pKa = 11.84AQSAEE162 pKa = 3.67PQRR165 pKa = 11.84PKK167 pKa = 10.02QRR169 pKa = 11.84EE170 pKa = 3.83RR171 pKa = 11.84SRR173 pKa = 11.84SAPSRR178 pKa = 11.84AAPKK182 pKa = 9.68QHH184 pKa = 7.29AIPKK188 pKa = 9.73RR189 pKa = 11.84VLLKK193 pKa = 10.81GKK195 pKa = 8.28TISGTFGKK203 pKa = 10.44RR204 pKa = 11.84SPAKK208 pKa = 10.52ANVGSPDD215 pKa = 3.57TEE217 pKa = 4.07KK218 pKa = 10.41TGLGDD223 pKa = 3.84PRR225 pKa = 11.84LMALMRR231 pKa = 11.84FVPGTQEE238 pKa = 3.95LLFAGHH244 pKa = 7.68LKK246 pKa = 10.6HH247 pKa = 6.84KK248 pKa = 7.01MQPEE252 pKa = 4.91GVQLTFTYY260 pKa = 10.56DD261 pKa = 2.96ITVKK265 pKa = 10.4RR266 pKa = 11.84DD267 pKa = 3.12NKK269 pKa = 10.68DD270 pKa = 2.6FDD272 pKa = 5.24RR273 pKa = 11.84IVEE276 pKa = 4.13ALNSVVDD283 pKa = 3.63MVYY286 pKa = 10.43EE287 pKa = 3.89PEE289 pKa = 4.14GEE291 pKa = 3.95IRR293 pKa = 11.84KK294 pKa = 8.97DD295 pKa = 3.46QQSKK299 pKa = 11.15KK300 pKa = 9.59DD301 pKa = 3.58TQPKK305 pKa = 9.62KK306 pKa = 10.07KK307 pKa = 10.21AEE309 pKa = 4.3KK310 pKa = 8.91KK311 pKa = 9.19TPTNPQKK318 pKa = 10.9QPDD321 pKa = 4.11KK322 pKa = 10.48PVATIVDD329 pKa = 3.74NGDD332 pKa = 3.44SQVINWGTDD341 pKa = 3.19PAEE344 pKa = 4.29GLEE347 pKa = 4.34SFSS350 pKa = 4.03
Molecular weight: 39.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8473 |
66 |
6206 |
847.3 |
94.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.07 ± 0.297 | 2.655 ± 0.338 |
5.618 ± 0.469 | 3.446 ± 0.239 |
4.473 ± 0.368 | 5.512 ± 0.479 |
2.16 ± 0.379 | 5.996 ± 0.476 |
4.851 ± 0.95 | 9.465 ± 0.707 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.865 ± 0.103 | 5.417 ± 0.865 |
4.603 ± 0.966 | 3.836 ± 0.636 |
3.493 ± 0.525 | 6.763 ± 0.583 |
7.223 ± 0.687 | 9.418 ± 0.712 |
0.956 ± 0.287 | 5.181 ± 0.638 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
