
Hubei tombus-like virus 43
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.09
Get precalculated fractions of proteins
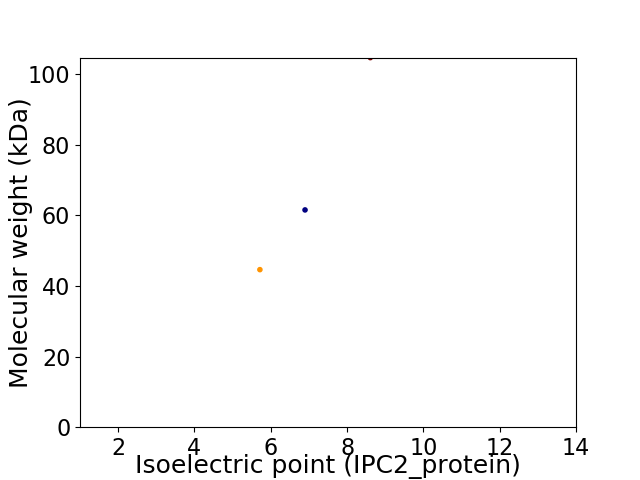
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGM0|A0A1L3KGM0_9VIRU Capsid protein OS=Hubei tombus-like virus 43 OX=1923291 PE=3 SV=1
MM1 pKa = 7.8PNKK4 pKa = 8.11QTKK7 pKa = 8.55RR8 pKa = 11.84TTKK11 pKa = 8.44NTKK14 pKa = 9.46PKK16 pKa = 8.15QQRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 9.03QPKK24 pKa = 7.37AQRR27 pKa = 11.84NVVSTQSAPVSRR39 pKa = 11.84GHH41 pKa = 7.29RR42 pKa = 11.84IKK44 pKa = 9.81QTAPKK49 pKa = 8.53ITASSKK55 pKa = 9.93GVNVVHH61 pKa = 6.79SEE63 pKa = 3.79ARR65 pKa = 11.84SVAVTSGSSTVDD77 pKa = 3.19MSWTLLPQGSNFPWLAAMAIQFDD100 pKa = 4.17KK101 pKa = 11.07IVWNKK106 pKa = 9.84VSVTFVPIVGTNLGGDD122 pKa = 3.29VVMYY126 pKa = 10.25FDD128 pKa = 4.96TEE130 pKa = 4.22ADD132 pKa = 3.61DD133 pKa = 4.08TRR135 pKa = 11.84VTDD138 pKa = 3.67YY139 pKa = 11.36VSAYY143 pKa = 10.22SMEE146 pKa = 4.27GAVSGRR152 pKa = 11.84VWDD155 pKa = 3.65MCTLHH160 pKa = 6.98AKK162 pKa = 10.01HH163 pKa = 6.95ADD165 pKa = 3.17LHH167 pKa = 5.27NQKK170 pKa = 10.48SYY172 pKa = 10.21AAKK175 pKa = 10.71GEE177 pKa = 4.31TSKK180 pKa = 11.46SLIGSPGMFCMLATLQTAATANTIIGRR207 pKa = 11.84WVINYY212 pKa = 8.69DD213 pKa = 3.48VTFLKK218 pKa = 10.7PEE220 pKa = 3.95AVTASSLLSLAGEE233 pKa = 4.78LDD235 pKa = 3.57PTAMRR240 pKa = 11.84TYY242 pKa = 10.54IEE244 pKa = 4.32AHH246 pKa = 4.65WTDD249 pKa = 4.52YY250 pKa = 8.8YY251 pKa = 10.9THH253 pKa = 7.19AACVSNCPCYY263 pKa = 10.16TSGDD267 pKa = 3.63QHH269 pKa = 5.22EE270 pKa = 4.41QCYY273 pKa = 10.4EE274 pKa = 3.79EE275 pKa = 4.84ALVGGMCLVDD285 pKa = 5.23KK286 pKa = 10.76VTDD289 pKa = 3.65TCSIMCPCNSLKK301 pKa = 10.6NASLGHH307 pKa = 5.62KK308 pKa = 10.17HH309 pKa = 6.1MIGHH313 pKa = 5.91SKK315 pKa = 10.81DD316 pKa = 3.8LIEE319 pKa = 4.4MDD321 pKa = 3.85KK322 pKa = 8.37TTHH325 pKa = 6.6DD326 pKa = 4.33AEE328 pKa = 4.31PMEE331 pKa = 4.53VDD333 pKa = 4.35DD334 pKa = 6.29LVAINQQPTLADD346 pKa = 3.58EE347 pKa = 4.28LTSRR351 pKa = 11.84GWVLNLPVDD360 pKa = 4.44NNDD363 pKa = 3.18EE364 pKa = 4.0MAKK367 pKa = 9.65FLKK370 pKa = 10.8AIDD373 pKa = 4.74IIDD376 pKa = 3.68KK377 pKa = 8.31VTSVRR382 pKa = 11.84EE383 pKa = 4.27GDD385 pKa = 3.05HH386 pKa = 6.83HH387 pKa = 6.2MWNGQTIHH395 pKa = 6.95ASSTSSSDD403 pKa = 3.5SEE405 pKa = 4.0MDD407 pKa = 3.08
MM1 pKa = 7.8PNKK4 pKa = 8.11QTKK7 pKa = 8.55RR8 pKa = 11.84TTKK11 pKa = 8.44NTKK14 pKa = 9.46PKK16 pKa = 8.15QQRR19 pKa = 11.84RR20 pKa = 11.84KK21 pKa = 9.03QPKK24 pKa = 7.37AQRR27 pKa = 11.84NVVSTQSAPVSRR39 pKa = 11.84GHH41 pKa = 7.29RR42 pKa = 11.84IKK44 pKa = 9.81QTAPKK49 pKa = 8.53ITASSKK55 pKa = 9.93GVNVVHH61 pKa = 6.79SEE63 pKa = 3.79ARR65 pKa = 11.84SVAVTSGSSTVDD77 pKa = 3.19MSWTLLPQGSNFPWLAAMAIQFDD100 pKa = 4.17KK101 pKa = 11.07IVWNKK106 pKa = 9.84VSVTFVPIVGTNLGGDD122 pKa = 3.29VVMYY126 pKa = 10.25FDD128 pKa = 4.96TEE130 pKa = 4.22ADD132 pKa = 3.61DD133 pKa = 4.08TRR135 pKa = 11.84VTDD138 pKa = 3.67YY139 pKa = 11.36VSAYY143 pKa = 10.22SMEE146 pKa = 4.27GAVSGRR152 pKa = 11.84VWDD155 pKa = 3.65MCTLHH160 pKa = 6.98AKK162 pKa = 10.01HH163 pKa = 6.95ADD165 pKa = 3.17LHH167 pKa = 5.27NQKK170 pKa = 10.48SYY172 pKa = 10.21AAKK175 pKa = 10.71GEE177 pKa = 4.31TSKK180 pKa = 11.46SLIGSPGMFCMLATLQTAATANTIIGRR207 pKa = 11.84WVINYY212 pKa = 8.69DD213 pKa = 3.48VTFLKK218 pKa = 10.7PEE220 pKa = 3.95AVTASSLLSLAGEE233 pKa = 4.78LDD235 pKa = 3.57PTAMRR240 pKa = 11.84TYY242 pKa = 10.54IEE244 pKa = 4.32AHH246 pKa = 4.65WTDD249 pKa = 4.52YY250 pKa = 8.8YY251 pKa = 10.9THH253 pKa = 7.19AACVSNCPCYY263 pKa = 10.16TSGDD267 pKa = 3.63QHH269 pKa = 5.22EE270 pKa = 4.41QCYY273 pKa = 10.4EE274 pKa = 3.79EE275 pKa = 4.84ALVGGMCLVDD285 pKa = 5.23KK286 pKa = 10.76VTDD289 pKa = 3.65TCSIMCPCNSLKK301 pKa = 10.6NASLGHH307 pKa = 5.62KK308 pKa = 10.17HH309 pKa = 6.1MIGHH313 pKa = 5.91SKK315 pKa = 10.81DD316 pKa = 3.8LIEE319 pKa = 4.4MDD321 pKa = 3.85KK322 pKa = 8.37TTHH325 pKa = 6.6DD326 pKa = 4.33AEE328 pKa = 4.31PMEE331 pKa = 4.53VDD333 pKa = 4.35DD334 pKa = 6.29LVAINQQPTLADD346 pKa = 3.58EE347 pKa = 4.28LTSRR351 pKa = 11.84GWVLNLPVDD360 pKa = 4.44NNDD363 pKa = 3.18EE364 pKa = 4.0MAKK367 pKa = 9.65FLKK370 pKa = 10.8AIDD373 pKa = 4.74IIDD376 pKa = 3.68KK377 pKa = 8.31VTSVRR382 pKa = 11.84EE383 pKa = 4.27GDD385 pKa = 3.05HH386 pKa = 6.83HH387 pKa = 6.2MWNGQTIHH395 pKa = 6.95ASSTSSSDD403 pKa = 3.5SEE405 pKa = 4.0MDD407 pKa = 3.08
Molecular weight: 44.64 kDa
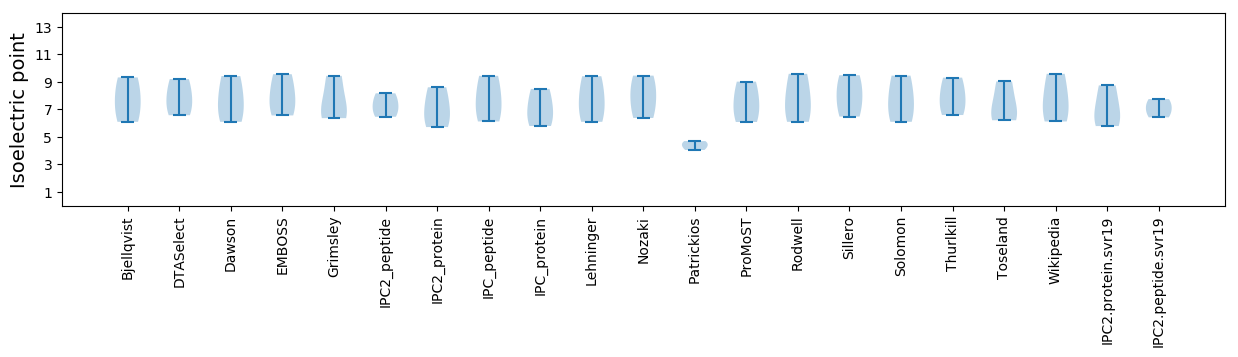
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGM0|A0A1L3KGM0_9VIRU Capsid protein OS=Hubei tombus-like virus 43 OX=1923291 PE=3 SV=1
MM1 pKa = 7.73NIFTPARR8 pKa = 11.84FFFKK12 pKa = 9.75ITCSTKK18 pKa = 10.05KK19 pKa = 9.83RR20 pKa = 11.84ALVSFLVLAASLAAFVGFLILLLAFQHH47 pKa = 5.73VDD49 pKa = 3.26YY50 pKa = 11.52VPIVDD55 pKa = 3.78VNSARR60 pKa = 11.84WIEE63 pKa = 4.14QQRR66 pKa = 11.84VTYY69 pKa = 10.3AIRR72 pKa = 11.84SYY74 pKa = 11.53SLIEE78 pKa = 3.93SLVILFTSILNRR90 pKa = 11.84LDD92 pKa = 3.39LTINMQPAIHH102 pKa = 6.57ILSPTMHH109 pKa = 6.22QMIITQRR116 pKa = 11.84EE117 pKa = 3.8QCDD120 pKa = 3.66YY121 pKa = 10.48WDD123 pKa = 3.61TALRR127 pKa = 11.84FYY129 pKa = 11.05DD130 pKa = 3.51EE131 pKa = 4.83HH132 pKa = 7.86LKK134 pKa = 10.69QFDD137 pKa = 3.78DD138 pKa = 4.02AGSYY142 pKa = 10.85SAMFVLTGTQIAGLAHH158 pKa = 6.38SEE160 pKa = 4.21SNIYY164 pKa = 10.29CRR166 pKa = 11.84GLSRR170 pKa = 11.84LGYY173 pKa = 10.22ASLPRR178 pKa = 11.84AYY180 pKa = 9.28PKK182 pKa = 10.42SWLRR186 pKa = 11.84RR187 pKa = 11.84IDD189 pKa = 4.65LINSEE194 pKa = 3.76QRR196 pKa = 11.84FYY198 pKa = 11.91NKK200 pKa = 9.55IAMRR204 pKa = 11.84LDD206 pKa = 3.89MIYY209 pKa = 10.5QLADD213 pKa = 4.08EE214 pKa = 5.38IIVGVLDD221 pKa = 4.11NIMWVLRR228 pKa = 11.84HH229 pKa = 6.21PFLSTFVVLLGLFVMMAIGFYY250 pKa = 10.73LSTLYY255 pKa = 10.49HH256 pKa = 5.9IQWSRR261 pKa = 11.84KK262 pKa = 8.34IKK264 pKa = 10.08NCAFAIRR271 pKa = 11.84QSAPLLPFRR280 pKa = 11.84PGVVKK285 pKa = 10.71DD286 pKa = 3.38VLKK289 pKa = 10.94RR290 pKa = 11.84RR291 pKa = 11.84LKK293 pKa = 9.41PLKK296 pKa = 9.21VTPTPDD302 pKa = 2.61GHH304 pKa = 8.45LCMAEE309 pKa = 3.84VRR311 pKa = 11.84RR312 pKa = 11.84NMEE315 pKa = 3.78RR316 pKa = 11.84TCIGFLLEE324 pKa = 3.97YY325 pKa = 9.75FKK327 pKa = 10.59QVRR330 pKa = 11.84DD331 pKa = 3.39IGGSPTRR338 pKa = 11.84NSHH341 pKa = 4.75YY342 pKa = 10.5GRR344 pKa = 11.84RR345 pKa = 11.84LHH347 pKa = 6.69ICCPDD352 pKa = 3.74LTAIDD357 pKa = 3.87HH358 pKa = 6.95DD359 pKa = 4.17KK360 pKa = 11.63VNIANYY366 pKa = 10.04KK367 pKa = 10.56NDD369 pKa = 3.79VQCHH373 pKa = 5.08KK374 pKa = 11.15SIDD377 pKa = 3.93CNVAKK382 pKa = 10.14QIPAAIMSYY391 pKa = 11.04VDD393 pKa = 4.52FHH395 pKa = 8.76IPIHH399 pKa = 6.48EE400 pKa = 4.47IADD403 pKa = 3.95SVTGPTLVITHH414 pKa = 6.44EE415 pKa = 4.58FKK417 pKa = 11.03EE418 pKa = 4.56GMDD421 pKa = 3.77EE422 pKa = 3.87KK423 pKa = 10.65WFDD426 pKa = 3.82GEE428 pKa = 4.59CTVKK432 pKa = 10.95ASGGLVQMSVRR443 pKa = 11.84GGNEE447 pKa = 3.41YY448 pKa = 10.67SHH450 pKa = 7.02YY451 pKa = 9.18YY452 pKa = 9.91HH453 pKa = 7.47DD454 pKa = 4.11WKK456 pKa = 11.23DD457 pKa = 3.32EE458 pKa = 3.79GVIIGKK464 pKa = 8.38SHH466 pKa = 7.35AINYY470 pKa = 9.25IKK472 pKa = 9.91VAKK475 pKa = 9.01YY476 pKa = 8.09EE477 pKa = 3.92HH478 pKa = 6.99SIVILLWPVTGTFTRR493 pKa = 11.84YY494 pKa = 10.19DD495 pKa = 3.61PNALFDD501 pKa = 3.93LRR503 pKa = 11.84YY504 pKa = 10.58DD505 pKa = 4.31EE506 pKa = 5.68ISLPDD511 pKa = 3.7GSSVVNTGKK520 pKa = 9.96HH521 pKa = 5.58FEE523 pKa = 4.41VKK525 pKa = 10.67DD526 pKa = 3.59NMGGHH531 pKa = 7.47LGLIPVDD538 pKa = 3.87TIVRR542 pKa = 11.84TAFTAKK548 pKa = 9.95GMARR552 pKa = 11.84DD553 pKa = 3.78PKK555 pKa = 10.84YY556 pKa = 10.61VANVDD561 pKa = 3.16ALLRR565 pKa = 11.84ARR567 pKa = 11.84FTQDD571 pKa = 2.59KK572 pKa = 10.4CDD574 pKa = 3.56MSLLPQASLVTYY586 pKa = 10.41RR587 pKa = 11.84LADD590 pKa = 4.39DD591 pKa = 4.34YY592 pKa = 11.47SLTMNHH598 pKa = 6.02MSSVIDD604 pKa = 4.09GPVVGLTTMQRR615 pKa = 11.84LRR617 pKa = 11.84CATLTLIKK625 pKa = 10.22GWTPRR630 pKa = 11.84RR631 pKa = 11.84FMHH634 pKa = 7.05LYY636 pKa = 10.46ASLLRR641 pKa = 11.84WCIGSDD647 pKa = 3.79QDD649 pKa = 5.26SSWLPWLWTDD659 pKa = 4.14HH660 pKa = 6.01VAPNYY665 pKa = 9.64EE666 pKa = 4.42VYY668 pKa = 10.26TPGSLSLMEE677 pKa = 4.51IKK679 pKa = 10.33QNKK682 pKa = 8.21QMLNAQPFQLPGQSCSTHH700 pKa = 5.71SAPNNEE706 pKa = 4.34CAAVKK711 pKa = 9.92HH712 pKa = 5.09VRR714 pKa = 11.84KK715 pKa = 8.55PTVKK719 pKa = 9.84RR720 pKa = 11.84VKK722 pKa = 10.35EE723 pKa = 4.09STKK726 pKa = 10.76DD727 pKa = 3.26QTPATSSTHH736 pKa = 6.25RR737 pKa = 11.84KK738 pKa = 7.98PSRR741 pKa = 11.84GEE743 pKa = 3.82RR744 pKa = 11.84KK745 pKa = 9.45NDD747 pKa = 2.88RR748 pKa = 11.84SAAAVAPEE756 pKa = 3.95PTQRR760 pKa = 11.84LGPSRR765 pKa = 11.84RR766 pKa = 11.84IPDD769 pKa = 3.3RR770 pKa = 11.84AVPNAGRR777 pKa = 11.84RR778 pKa = 11.84HH779 pKa = 4.42QQSGGRR785 pKa = 11.84NDD787 pKa = 4.18ASGKK791 pKa = 8.18STADD795 pKa = 3.3RR796 pKa = 11.84RR797 pKa = 11.84PDD799 pKa = 3.01RR800 pKa = 11.84RR801 pKa = 11.84NADD804 pKa = 3.32KK805 pKa = 11.36VNTRR809 pKa = 11.84TTPALNDD816 pKa = 3.07RR817 pKa = 11.84GMGKK821 pKa = 9.71KK822 pKa = 10.36VSGKK826 pKa = 8.3STSRR830 pKa = 11.84PNSSPRR836 pKa = 11.84KK837 pKa = 8.03SKK839 pKa = 10.95SVVHH843 pKa = 6.6AKK845 pKa = 9.89EE846 pKa = 3.81LQQRR850 pKa = 11.84PNIPEE855 pKa = 3.44IRR857 pKa = 11.84NVDD860 pKa = 3.35KK861 pKa = 11.35SDD863 pKa = 3.33GPKK866 pKa = 9.68EE867 pKa = 4.08HH868 pKa = 6.75NPEE871 pKa = 3.99STRR874 pKa = 11.84VPFDD878 pKa = 3.25FRR880 pKa = 11.84AFCEE884 pKa = 4.16QYY886 pKa = 10.33RR887 pKa = 11.84EE888 pKa = 4.47CNPPPPIPCEE898 pKa = 3.83RR899 pKa = 11.84QEE901 pKa = 4.15QLGPRR906 pKa = 11.84SEE908 pKa = 4.88NIPEE912 pKa = 4.17IKK914 pKa = 10.02QLPSVCGDD922 pKa = 3.72GLQQ925 pKa = 3.7
MM1 pKa = 7.73NIFTPARR8 pKa = 11.84FFFKK12 pKa = 9.75ITCSTKK18 pKa = 10.05KK19 pKa = 9.83RR20 pKa = 11.84ALVSFLVLAASLAAFVGFLILLLAFQHH47 pKa = 5.73VDD49 pKa = 3.26YY50 pKa = 11.52VPIVDD55 pKa = 3.78VNSARR60 pKa = 11.84WIEE63 pKa = 4.14QQRR66 pKa = 11.84VTYY69 pKa = 10.3AIRR72 pKa = 11.84SYY74 pKa = 11.53SLIEE78 pKa = 3.93SLVILFTSILNRR90 pKa = 11.84LDD92 pKa = 3.39LTINMQPAIHH102 pKa = 6.57ILSPTMHH109 pKa = 6.22QMIITQRR116 pKa = 11.84EE117 pKa = 3.8QCDD120 pKa = 3.66YY121 pKa = 10.48WDD123 pKa = 3.61TALRR127 pKa = 11.84FYY129 pKa = 11.05DD130 pKa = 3.51EE131 pKa = 4.83HH132 pKa = 7.86LKK134 pKa = 10.69QFDD137 pKa = 3.78DD138 pKa = 4.02AGSYY142 pKa = 10.85SAMFVLTGTQIAGLAHH158 pKa = 6.38SEE160 pKa = 4.21SNIYY164 pKa = 10.29CRR166 pKa = 11.84GLSRR170 pKa = 11.84LGYY173 pKa = 10.22ASLPRR178 pKa = 11.84AYY180 pKa = 9.28PKK182 pKa = 10.42SWLRR186 pKa = 11.84RR187 pKa = 11.84IDD189 pKa = 4.65LINSEE194 pKa = 3.76QRR196 pKa = 11.84FYY198 pKa = 11.91NKK200 pKa = 9.55IAMRR204 pKa = 11.84LDD206 pKa = 3.89MIYY209 pKa = 10.5QLADD213 pKa = 4.08EE214 pKa = 5.38IIVGVLDD221 pKa = 4.11NIMWVLRR228 pKa = 11.84HH229 pKa = 6.21PFLSTFVVLLGLFVMMAIGFYY250 pKa = 10.73LSTLYY255 pKa = 10.49HH256 pKa = 5.9IQWSRR261 pKa = 11.84KK262 pKa = 8.34IKK264 pKa = 10.08NCAFAIRR271 pKa = 11.84QSAPLLPFRR280 pKa = 11.84PGVVKK285 pKa = 10.71DD286 pKa = 3.38VLKK289 pKa = 10.94RR290 pKa = 11.84RR291 pKa = 11.84LKK293 pKa = 9.41PLKK296 pKa = 9.21VTPTPDD302 pKa = 2.61GHH304 pKa = 8.45LCMAEE309 pKa = 3.84VRR311 pKa = 11.84RR312 pKa = 11.84NMEE315 pKa = 3.78RR316 pKa = 11.84TCIGFLLEE324 pKa = 3.97YY325 pKa = 9.75FKK327 pKa = 10.59QVRR330 pKa = 11.84DD331 pKa = 3.39IGGSPTRR338 pKa = 11.84NSHH341 pKa = 4.75YY342 pKa = 10.5GRR344 pKa = 11.84RR345 pKa = 11.84LHH347 pKa = 6.69ICCPDD352 pKa = 3.74LTAIDD357 pKa = 3.87HH358 pKa = 6.95DD359 pKa = 4.17KK360 pKa = 11.63VNIANYY366 pKa = 10.04KK367 pKa = 10.56NDD369 pKa = 3.79VQCHH373 pKa = 5.08KK374 pKa = 11.15SIDD377 pKa = 3.93CNVAKK382 pKa = 10.14QIPAAIMSYY391 pKa = 11.04VDD393 pKa = 4.52FHH395 pKa = 8.76IPIHH399 pKa = 6.48EE400 pKa = 4.47IADD403 pKa = 3.95SVTGPTLVITHH414 pKa = 6.44EE415 pKa = 4.58FKK417 pKa = 11.03EE418 pKa = 4.56GMDD421 pKa = 3.77EE422 pKa = 3.87KK423 pKa = 10.65WFDD426 pKa = 3.82GEE428 pKa = 4.59CTVKK432 pKa = 10.95ASGGLVQMSVRR443 pKa = 11.84GGNEE447 pKa = 3.41YY448 pKa = 10.67SHH450 pKa = 7.02YY451 pKa = 9.18YY452 pKa = 9.91HH453 pKa = 7.47DD454 pKa = 4.11WKK456 pKa = 11.23DD457 pKa = 3.32EE458 pKa = 3.79GVIIGKK464 pKa = 8.38SHH466 pKa = 7.35AINYY470 pKa = 9.25IKK472 pKa = 9.91VAKK475 pKa = 9.01YY476 pKa = 8.09EE477 pKa = 3.92HH478 pKa = 6.99SIVILLWPVTGTFTRR493 pKa = 11.84YY494 pKa = 10.19DD495 pKa = 3.61PNALFDD501 pKa = 3.93LRR503 pKa = 11.84YY504 pKa = 10.58DD505 pKa = 4.31EE506 pKa = 5.68ISLPDD511 pKa = 3.7GSSVVNTGKK520 pKa = 9.96HH521 pKa = 5.58FEE523 pKa = 4.41VKK525 pKa = 10.67DD526 pKa = 3.59NMGGHH531 pKa = 7.47LGLIPVDD538 pKa = 3.87TIVRR542 pKa = 11.84TAFTAKK548 pKa = 9.95GMARR552 pKa = 11.84DD553 pKa = 3.78PKK555 pKa = 10.84YY556 pKa = 10.61VANVDD561 pKa = 3.16ALLRR565 pKa = 11.84ARR567 pKa = 11.84FTQDD571 pKa = 2.59KK572 pKa = 10.4CDD574 pKa = 3.56MSLLPQASLVTYY586 pKa = 10.41RR587 pKa = 11.84LADD590 pKa = 4.39DD591 pKa = 4.34YY592 pKa = 11.47SLTMNHH598 pKa = 6.02MSSVIDD604 pKa = 4.09GPVVGLTTMQRR615 pKa = 11.84LRR617 pKa = 11.84CATLTLIKK625 pKa = 10.22GWTPRR630 pKa = 11.84RR631 pKa = 11.84FMHH634 pKa = 7.05LYY636 pKa = 10.46ASLLRR641 pKa = 11.84WCIGSDD647 pKa = 3.79QDD649 pKa = 5.26SSWLPWLWTDD659 pKa = 4.14HH660 pKa = 6.01VAPNYY665 pKa = 9.64EE666 pKa = 4.42VYY668 pKa = 10.26TPGSLSLMEE677 pKa = 4.51IKK679 pKa = 10.33QNKK682 pKa = 8.21QMLNAQPFQLPGQSCSTHH700 pKa = 5.71SAPNNEE706 pKa = 4.34CAAVKK711 pKa = 9.92HH712 pKa = 5.09VRR714 pKa = 11.84KK715 pKa = 8.55PTVKK719 pKa = 9.84RR720 pKa = 11.84VKK722 pKa = 10.35EE723 pKa = 4.09STKK726 pKa = 10.76DD727 pKa = 3.26QTPATSSTHH736 pKa = 6.25RR737 pKa = 11.84KK738 pKa = 7.98PSRR741 pKa = 11.84GEE743 pKa = 3.82RR744 pKa = 11.84KK745 pKa = 9.45NDD747 pKa = 2.88RR748 pKa = 11.84SAAAVAPEE756 pKa = 3.95PTQRR760 pKa = 11.84LGPSRR765 pKa = 11.84RR766 pKa = 11.84IPDD769 pKa = 3.3RR770 pKa = 11.84AVPNAGRR777 pKa = 11.84RR778 pKa = 11.84HH779 pKa = 4.42QQSGGRR785 pKa = 11.84NDD787 pKa = 4.18ASGKK791 pKa = 8.18STADD795 pKa = 3.3RR796 pKa = 11.84RR797 pKa = 11.84PDD799 pKa = 3.01RR800 pKa = 11.84RR801 pKa = 11.84NADD804 pKa = 3.32KK805 pKa = 11.36VNTRR809 pKa = 11.84TTPALNDD816 pKa = 3.07RR817 pKa = 11.84GMGKK821 pKa = 9.71KK822 pKa = 10.36VSGKK826 pKa = 8.3STSRR830 pKa = 11.84PNSSPRR836 pKa = 11.84KK837 pKa = 8.03SKK839 pKa = 10.95SVVHH843 pKa = 6.6AKK845 pKa = 9.89EE846 pKa = 3.81LQQRR850 pKa = 11.84PNIPEE855 pKa = 3.44IRR857 pKa = 11.84NVDD860 pKa = 3.35KK861 pKa = 11.35SDD863 pKa = 3.33GPKK866 pKa = 9.68EE867 pKa = 4.08HH868 pKa = 6.75NPEE871 pKa = 3.99STRR874 pKa = 11.84VPFDD878 pKa = 3.25FRR880 pKa = 11.84AFCEE884 pKa = 4.16QYY886 pKa = 10.33RR887 pKa = 11.84EE888 pKa = 4.47CNPPPPIPCEE898 pKa = 3.83RR899 pKa = 11.84QEE901 pKa = 4.15QLGPRR906 pKa = 11.84SEE908 pKa = 4.88NIPEE912 pKa = 4.17IKK914 pKa = 10.02QLPSVCGDD922 pKa = 3.72GLQQ925 pKa = 3.7
Molecular weight: 104.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1874 |
407 |
925 |
624.7 |
70.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.937 ± 0.548 | 2.081 ± 0.167 |
5.923 ± 0.323 | 4.269 ± 0.273 |
3.308 ± 0.515 | 4.909 ± 0.446 |
3.308 ± 0.123 | 6.03 ± 0.528 |
5.39 ± 0.275 | 8.645 ± 0.932 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.775 ± 0.485 | 4.482 ± 0.192 |
5.923 ± 0.662 | 4.055 ± 0.07 |
6.03 ± 1.064 | 7.737 ± 0.442 |
6.724 ± 0.858 | 6.83 ± 0.416 |
1.708 ± 0.186 | 2.935 ± 0.261 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
