
Oryza nivara (Indian wild rice) (Oryza sativa f. spontanea)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; BOP clade; Oryzoideae; Oryzeae;
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
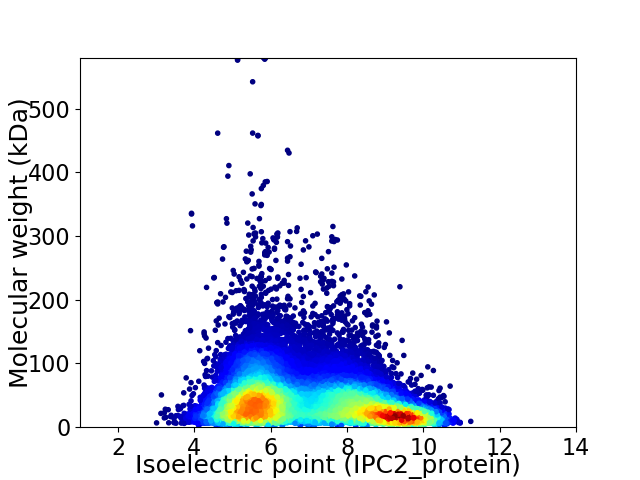
Virtual 2D-PAGE plot for 46176 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E0IHB6|A0A0E0IHB6_ORYNI Isoform of A0A0E0IHB5 Uncharacterized protein OS=Oryza nivara OX=4536 PE=4 SV=1
MM1 pKa = 7.95DD2 pKa = 4.96GVDD5 pKa = 5.1DD6 pKa = 5.24GLICSTSCYY15 pKa = 10.16KK16 pKa = 10.0MCGHH20 pKa = 7.43ADD22 pKa = 3.17KK23 pKa = 11.3SSIIDD28 pKa = 4.26GEE30 pKa = 4.3IGWPSMEE37 pKa = 4.71ISPCSTPYY45 pKa = 9.15GTPLFSRR52 pKa = 11.84EE53 pKa = 4.13SSCSSFASCFSSLDD67 pKa = 3.68DD68 pKa = 4.04YY69 pKa = 11.74LVEE72 pKa = 4.24TDD74 pKa = 3.63SEE76 pKa = 4.47EE77 pKa = 4.28EE78 pKa = 4.15IEE80 pKa = 5.05VLDD83 pKa = 4.09TGQLHH88 pKa = 6.57PGILFSDD95 pKa = 3.83EE96 pKa = 4.47SMEE99 pKa = 4.07QRR101 pKa = 11.84EE102 pKa = 4.13ADD104 pKa = 3.44SVQVEE109 pKa = 5.15EE110 pKa = 4.52YY111 pKa = 9.96QVSHH115 pKa = 6.15SAVVNDD121 pKa = 4.95DD122 pKa = 3.64SSISIPTNQNNSSGQLQLEE141 pKa = 4.18IHH143 pKa = 6.92LDD145 pKa = 3.38ATNEE149 pKa = 3.79KK150 pKa = 9.98SVPSNAILDD159 pKa = 4.05ANVTDD164 pKa = 3.78PHH166 pKa = 7.84QEE168 pKa = 4.02VISNGGLIEE177 pKa = 4.19ACYY180 pKa = 9.82GVPVDD185 pKa = 6.53DD186 pKa = 5.97IDD188 pKa = 5.13LKK190 pKa = 11.05QSNILDD196 pKa = 3.97GEE198 pKa = 4.73EE199 pKa = 3.99ITSLPMADD207 pKa = 4.82NEE209 pKa = 4.34MTPLDD214 pKa = 3.98DD215 pKa = 3.7MKK217 pKa = 10.5EE218 pKa = 3.85ISSIVYY224 pKa = 9.89NSTISAEE231 pKa = 3.89QHH233 pKa = 5.68VNSGSEE239 pKa = 4.02FEE241 pKa = 4.85KK242 pKa = 11.14GNEE245 pKa = 4.27SSDD248 pKa = 3.31KK249 pKa = 10.67LYY251 pKa = 10.45PLVMPSFDD259 pKa = 3.98TDD261 pKa = 3.19PHH263 pKa = 6.55IWLPPDD269 pKa = 4.12PVNKK273 pKa = 10.43DD274 pKa = 3.22DD275 pKa = 5.16DD276 pKa = 4.13TDD278 pKa = 3.52IVANNDD284 pKa = 3.61DD285 pKa = 3.93NSDD288 pKa = 3.79NNGNWVQSSFNISFDD303 pKa = 3.09AKK305 pKa = 10.42RR306 pKa = 11.84NKK308 pKa = 9.02TSCEE312 pKa = 3.94DD313 pKa = 3.24QQ314 pKa = 3.61
MM1 pKa = 7.95DD2 pKa = 4.96GVDD5 pKa = 5.1DD6 pKa = 5.24GLICSTSCYY15 pKa = 10.16KK16 pKa = 10.0MCGHH20 pKa = 7.43ADD22 pKa = 3.17KK23 pKa = 11.3SSIIDD28 pKa = 4.26GEE30 pKa = 4.3IGWPSMEE37 pKa = 4.71ISPCSTPYY45 pKa = 9.15GTPLFSRR52 pKa = 11.84EE53 pKa = 4.13SSCSSFASCFSSLDD67 pKa = 3.68DD68 pKa = 4.04YY69 pKa = 11.74LVEE72 pKa = 4.24TDD74 pKa = 3.63SEE76 pKa = 4.47EE77 pKa = 4.28EE78 pKa = 4.15IEE80 pKa = 5.05VLDD83 pKa = 4.09TGQLHH88 pKa = 6.57PGILFSDD95 pKa = 3.83EE96 pKa = 4.47SMEE99 pKa = 4.07QRR101 pKa = 11.84EE102 pKa = 4.13ADD104 pKa = 3.44SVQVEE109 pKa = 5.15EE110 pKa = 4.52YY111 pKa = 9.96QVSHH115 pKa = 6.15SAVVNDD121 pKa = 4.95DD122 pKa = 3.64SSISIPTNQNNSSGQLQLEE141 pKa = 4.18IHH143 pKa = 6.92LDD145 pKa = 3.38ATNEE149 pKa = 3.79KK150 pKa = 9.98SVPSNAILDD159 pKa = 4.05ANVTDD164 pKa = 3.78PHH166 pKa = 7.84QEE168 pKa = 4.02VISNGGLIEE177 pKa = 4.19ACYY180 pKa = 9.82GVPVDD185 pKa = 6.53DD186 pKa = 5.97IDD188 pKa = 5.13LKK190 pKa = 11.05QSNILDD196 pKa = 3.97GEE198 pKa = 4.73EE199 pKa = 3.99ITSLPMADD207 pKa = 4.82NEE209 pKa = 4.34MTPLDD214 pKa = 3.98DD215 pKa = 3.7MKK217 pKa = 10.5EE218 pKa = 3.85ISSIVYY224 pKa = 9.89NSTISAEE231 pKa = 3.89QHH233 pKa = 5.68VNSGSEE239 pKa = 4.02FEE241 pKa = 4.85KK242 pKa = 11.14GNEE245 pKa = 4.27SSDD248 pKa = 3.31KK249 pKa = 10.67LYY251 pKa = 10.45PLVMPSFDD259 pKa = 3.98TDD261 pKa = 3.19PHH263 pKa = 6.55IWLPPDD269 pKa = 4.12PVNKK273 pKa = 10.43DD274 pKa = 3.22DD275 pKa = 5.16DD276 pKa = 4.13TDD278 pKa = 3.52IVANNDD284 pKa = 3.61DD285 pKa = 3.93NSDD288 pKa = 3.79NNGNWVQSSFNISFDD303 pKa = 3.09AKK305 pKa = 10.42RR306 pKa = 11.84NKK308 pKa = 9.02TSCEE312 pKa = 3.94DD313 pKa = 3.24QQ314 pKa = 3.61
Molecular weight: 34.3 kDa
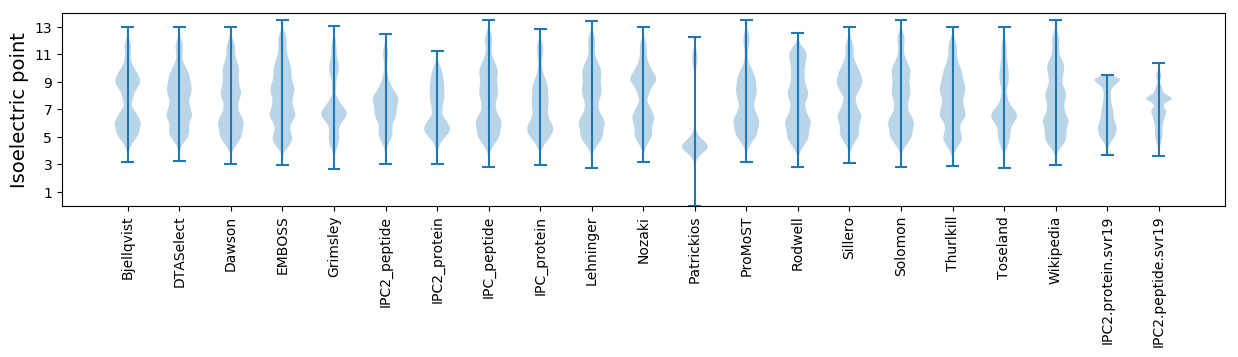
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E0J0V6|A0A0E0J0V6_ORYNI Ribonuclease P/MRP protein subunit POP5 OS=Oryza nivara OX=4536 PE=3 SV=1
MM1 pKa = 7.83PSLTTGRR8 pKa = 11.84SPWRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 10.25RR16 pKa = 11.84GTATTARR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84TPTSLRR32 pKa = 11.84ALWRR36 pKa = 11.84RR37 pKa = 11.84IVPRR41 pKa = 11.84TSTTTAPRR49 pKa = 11.84VRR51 pKa = 11.84TRR53 pKa = 11.84KK54 pKa = 9.6PGLLSRR60 pKa = 11.84ALRR63 pKa = 11.84VLSCGGGRR71 pKa = 11.84RR72 pKa = 11.84SRR74 pKa = 11.84AARR77 pKa = 11.84RR78 pKa = 11.84WW79 pKa = 3.29
MM1 pKa = 7.83PSLTTGRR8 pKa = 11.84SPWRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84KK15 pKa = 10.25RR16 pKa = 11.84GTATTARR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84TPTSLRR32 pKa = 11.84ALWRR36 pKa = 11.84RR37 pKa = 11.84IVPRR41 pKa = 11.84TSTTTAPRR49 pKa = 11.84VRR51 pKa = 11.84TRR53 pKa = 11.84KK54 pKa = 9.6PGLLSRR60 pKa = 11.84ALRR63 pKa = 11.84VLSCGGGRR71 pKa = 11.84RR72 pKa = 11.84SRR74 pKa = 11.84AARR77 pKa = 11.84RR78 pKa = 11.84WW79 pKa = 3.29
Molecular weight: 9.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
19239447 |
7 |
5485 |
416.7 |
45.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.148 ± 0.018 | 1.944 ± 0.005 |
5.372 ± 0.009 | 5.962 ± 0.013 |
3.62 ± 0.007 | 7.408 ± 0.014 |
2.516 ± 0.006 | 4.494 ± 0.009 |
4.833 ± 0.011 | 9.587 ± 0.017 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.432 ± 0.004 | 3.561 ± 0.01 |
5.545 ± 0.012 | 3.401 ± 0.008 |
6.419 ± 0.012 | 8.395 ± 0.011 |
4.809 ± 0.007 | 6.701 ± 0.009 |
1.319 ± 0.004 | 2.535 ± 0.006 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
