
Trematomus pennellii polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae
Average proteome isoelectric point is 5.81
Get precalculated fractions of proteins
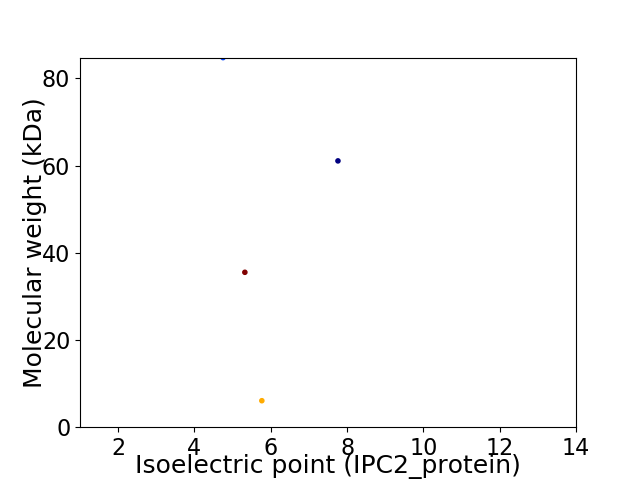
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
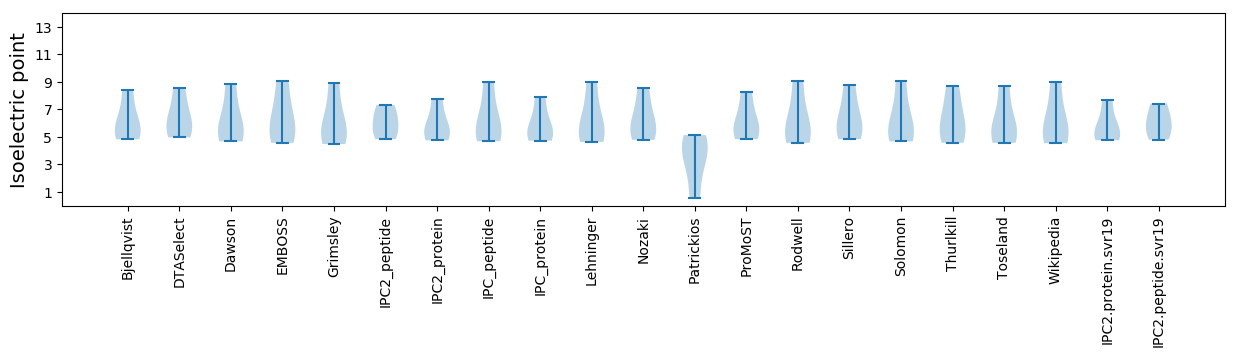
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0E3JRJ3|A0A0E3JRJ3_9POLY Large T-antigen OS=Trematomus pennellii polyomavirus 1 OX=2170103 PE=4 SV=1
MM1 pKa = 6.24QQEE4 pKa = 4.18YY5 pKa = 8.21TADD8 pKa = 3.36ANGRR12 pKa = 11.84NRR14 pKa = 11.84NLLEE18 pKa = 4.16TNKK21 pKa = 9.54EE22 pKa = 3.71VAMANSTTFTSNVYY36 pKa = 10.04ASKK39 pKa = 10.7LIKK42 pKa = 10.6ALGSSVFPGATMEE55 pKa = 5.32DD56 pKa = 4.82NMITWNADD64 pKa = 2.42MDD66 pKa = 4.26ALSFQDD72 pKa = 3.41SQGNKK77 pKa = 7.89EE78 pKa = 3.98DD79 pKa = 4.99RR80 pKa = 11.84ILLRR84 pKa = 11.84EE85 pKa = 4.13KK86 pKa = 11.25VNDD89 pKa = 3.19LWKK92 pKa = 10.52GYY94 pKa = 8.89QAAEE98 pKa = 3.77AVLADD103 pKa = 4.86FAPSDD108 pKa = 4.03TPKK111 pKa = 10.15MAGPVDD117 pKa = 4.35FCADD121 pKa = 3.2VTEE124 pKa = 5.59FLDD127 pKa = 4.2PDD129 pKa = 3.98CTSASAYY136 pKa = 10.94LFMAPASLYY145 pKa = 8.2PQLMRR150 pKa = 11.84IAKK153 pKa = 9.97GYY155 pKa = 10.55RR156 pKa = 11.84LDD158 pKa = 4.43DD159 pKa = 4.08YY160 pKa = 11.45YY161 pKa = 11.77GVAHH165 pKa = 6.81TDD167 pKa = 3.65EE168 pKa = 4.56KK169 pKa = 10.55WGEE172 pKa = 3.59IQYY175 pKa = 10.81FGFKK179 pKa = 8.83TRR181 pKa = 11.84YY182 pKa = 8.84RR183 pKa = 11.84LARR186 pKa = 11.84HH187 pKa = 4.73NMLRR191 pKa = 11.84KK192 pKa = 9.33MEE194 pKa = 4.46ASVSNKK200 pKa = 9.08GWVKK204 pKa = 10.23GCPVVSKK211 pKa = 10.35KK212 pKa = 10.66FNMCKK217 pKa = 9.92SHH219 pKa = 7.14LFNCAGAEE227 pKa = 4.16VTEE230 pKa = 4.55GAVEE234 pKa = 4.23FDD236 pKa = 3.67VLCFNQQTLARR247 pKa = 11.84YY248 pKa = 9.37AKK250 pKa = 9.59DD251 pKa = 3.02TGTCDD256 pKa = 3.5PLLLMGDD263 pKa = 3.7YY264 pKa = 9.81MAFSGLVHH272 pKa = 7.37ACTQCHH278 pKa = 4.6TEE280 pKa = 3.72RR281 pKa = 11.84RR282 pKa = 11.84RR283 pKa = 11.84GGQCHH288 pKa = 4.91ATYY291 pKa = 10.34HH292 pKa = 6.72KK293 pKa = 10.17EE294 pKa = 3.85HH295 pKa = 6.44QGNAKK300 pKa = 10.36AFMMVSNKK308 pKa = 9.52KK309 pKa = 9.78SAATNACDD317 pKa = 3.56EE318 pKa = 4.47SCAVLRR324 pKa = 11.84RR325 pKa = 11.84ASKK328 pKa = 10.37LPRR331 pKa = 11.84YY332 pKa = 9.77DD333 pKa = 3.54RR334 pKa = 11.84LKK336 pKa = 11.29DD337 pKa = 3.57NMDD340 pKa = 3.0IAMKK344 pKa = 10.44VLEE347 pKa = 4.9EE348 pKa = 4.06SDD350 pKa = 3.24ITNSQVAAAMLLSEE364 pKa = 5.78LIPGSVCDD372 pKa = 5.66FFDD375 pKa = 3.6QVISALLKK383 pKa = 10.77AEE385 pKa = 4.19PKK387 pKa = 9.66KK388 pKa = 10.63QGFIFRR394 pKa = 11.84GPYY397 pKa = 8.32NTGKK401 pKa = 8.58STVAAAVMSLLGGSSLNVNSPRR423 pKa = 11.84EE424 pKa = 3.98RR425 pKa = 11.84LWVEE429 pKa = 4.21FGLAMDD435 pKa = 4.52RR436 pKa = 11.84FAVCFEE442 pKa = 4.39DD443 pKa = 3.75VQGIPSPGEE452 pKa = 3.45PCLPFGEE459 pKa = 4.93GWSNLDD465 pKa = 3.36HH466 pKa = 6.53FRR468 pKa = 11.84EE469 pKa = 4.17YY470 pKa = 11.27LDD472 pKa = 4.13GLFKK476 pKa = 11.1VGLEE480 pKa = 4.05KK481 pKa = 10.57KK482 pKa = 9.21HH483 pKa = 5.37QNKK486 pKa = 6.99VTQNFPPWIATTNNYY501 pKa = 9.77IIPEE505 pKa = 4.33SVLEE509 pKa = 4.07RR510 pKa = 11.84AVVIPFKK517 pKa = 10.9RR518 pKa = 11.84MSFDD522 pKa = 3.14VQKK525 pKa = 10.91YY526 pKa = 9.4IEE528 pKa = 4.71EE529 pKa = 4.16FDD531 pKa = 3.25VDD533 pKa = 4.75LRR535 pKa = 11.84FLSSGPCLFLLWLLEE550 pKa = 4.36DD551 pKa = 4.13SVDD554 pKa = 5.48DD555 pKa = 5.45IDD557 pKa = 6.66DD558 pKa = 4.0EE559 pKa = 4.63LLDD562 pKa = 3.86RR563 pKa = 11.84AVVLSLAAKK572 pKa = 7.55MIWNAEE578 pKa = 3.63MRR580 pKa = 11.84DD581 pKa = 4.24FIKK584 pKa = 10.73APPPDD589 pKa = 3.97VLTAVYY595 pKa = 9.89VAEE598 pKa = 5.22DD599 pKa = 3.36GGIRR603 pKa = 11.84CTTGPEE609 pKa = 4.08KK610 pKa = 10.82EE611 pKa = 4.14NTPFTVSKK619 pKa = 9.63VWRR622 pKa = 11.84AIEE625 pKa = 4.08KK626 pKa = 10.6EE627 pKa = 4.1EE628 pKa = 4.03EE629 pKa = 3.92QLPEE633 pKa = 5.1DD634 pKa = 3.67IHH636 pKa = 5.79WTDD639 pKa = 3.65SEE641 pKa = 4.39SDD643 pKa = 3.72SVKK646 pKa = 10.83SVLATPLPFTPPDD659 pKa = 4.41PGLGSSDD666 pKa = 3.8EE667 pKa = 4.29EE668 pKa = 4.47EE669 pKa = 4.29EE670 pKa = 4.35VPPRR674 pKa = 11.84QIPKK678 pKa = 9.77KK679 pKa = 9.29RR680 pKa = 11.84KK681 pKa = 9.48CPFIDD686 pKa = 3.89EE687 pKa = 4.74AATEE691 pKa = 4.22CADD694 pKa = 4.95DD695 pKa = 4.08EE696 pKa = 4.63QSDD699 pKa = 3.82SSEE702 pKa = 4.25PYY704 pKa = 9.54PVPFEE709 pKa = 4.27KK710 pKa = 10.62KK711 pKa = 8.87KK712 pKa = 10.8GPRR715 pKa = 11.84VWVVVRR721 pKa = 11.84EE722 pKa = 4.19DD723 pKa = 3.51GAIGDD728 pKa = 4.5IEE730 pKa = 4.8NGPAPPPDD738 pKa = 3.69EE739 pKa = 4.18QSTSGFTDD747 pKa = 3.49ASGTTDD753 pKa = 3.87KK754 pKa = 11.22DD755 pKa = 3.88DD756 pKa = 5.08PDD758 pKa = 4.06VEE760 pKa = 4.61TSS762 pKa = 3.02
MM1 pKa = 6.24QQEE4 pKa = 4.18YY5 pKa = 8.21TADD8 pKa = 3.36ANGRR12 pKa = 11.84NRR14 pKa = 11.84NLLEE18 pKa = 4.16TNKK21 pKa = 9.54EE22 pKa = 3.71VAMANSTTFTSNVYY36 pKa = 10.04ASKK39 pKa = 10.7LIKK42 pKa = 10.6ALGSSVFPGATMEE55 pKa = 5.32DD56 pKa = 4.82NMITWNADD64 pKa = 2.42MDD66 pKa = 4.26ALSFQDD72 pKa = 3.41SQGNKK77 pKa = 7.89EE78 pKa = 3.98DD79 pKa = 4.99RR80 pKa = 11.84ILLRR84 pKa = 11.84EE85 pKa = 4.13KK86 pKa = 11.25VNDD89 pKa = 3.19LWKK92 pKa = 10.52GYY94 pKa = 8.89QAAEE98 pKa = 3.77AVLADD103 pKa = 4.86FAPSDD108 pKa = 4.03TPKK111 pKa = 10.15MAGPVDD117 pKa = 4.35FCADD121 pKa = 3.2VTEE124 pKa = 5.59FLDD127 pKa = 4.2PDD129 pKa = 3.98CTSASAYY136 pKa = 10.94LFMAPASLYY145 pKa = 8.2PQLMRR150 pKa = 11.84IAKK153 pKa = 9.97GYY155 pKa = 10.55RR156 pKa = 11.84LDD158 pKa = 4.43DD159 pKa = 4.08YY160 pKa = 11.45YY161 pKa = 11.77GVAHH165 pKa = 6.81TDD167 pKa = 3.65EE168 pKa = 4.56KK169 pKa = 10.55WGEE172 pKa = 3.59IQYY175 pKa = 10.81FGFKK179 pKa = 8.83TRR181 pKa = 11.84YY182 pKa = 8.84RR183 pKa = 11.84LARR186 pKa = 11.84HH187 pKa = 4.73NMLRR191 pKa = 11.84KK192 pKa = 9.33MEE194 pKa = 4.46ASVSNKK200 pKa = 9.08GWVKK204 pKa = 10.23GCPVVSKK211 pKa = 10.35KK212 pKa = 10.66FNMCKK217 pKa = 9.92SHH219 pKa = 7.14LFNCAGAEE227 pKa = 4.16VTEE230 pKa = 4.55GAVEE234 pKa = 4.23FDD236 pKa = 3.67VLCFNQQTLARR247 pKa = 11.84YY248 pKa = 9.37AKK250 pKa = 9.59DD251 pKa = 3.02TGTCDD256 pKa = 3.5PLLLMGDD263 pKa = 3.7YY264 pKa = 9.81MAFSGLVHH272 pKa = 7.37ACTQCHH278 pKa = 4.6TEE280 pKa = 3.72RR281 pKa = 11.84RR282 pKa = 11.84RR283 pKa = 11.84GGQCHH288 pKa = 4.91ATYY291 pKa = 10.34HH292 pKa = 6.72KK293 pKa = 10.17EE294 pKa = 3.85HH295 pKa = 6.44QGNAKK300 pKa = 10.36AFMMVSNKK308 pKa = 9.52KK309 pKa = 9.78SAATNACDD317 pKa = 3.56EE318 pKa = 4.47SCAVLRR324 pKa = 11.84RR325 pKa = 11.84ASKK328 pKa = 10.37LPRR331 pKa = 11.84YY332 pKa = 9.77DD333 pKa = 3.54RR334 pKa = 11.84LKK336 pKa = 11.29DD337 pKa = 3.57NMDD340 pKa = 3.0IAMKK344 pKa = 10.44VLEE347 pKa = 4.9EE348 pKa = 4.06SDD350 pKa = 3.24ITNSQVAAAMLLSEE364 pKa = 5.78LIPGSVCDD372 pKa = 5.66FFDD375 pKa = 3.6QVISALLKK383 pKa = 10.77AEE385 pKa = 4.19PKK387 pKa = 9.66KK388 pKa = 10.63QGFIFRR394 pKa = 11.84GPYY397 pKa = 8.32NTGKK401 pKa = 8.58STVAAAVMSLLGGSSLNVNSPRR423 pKa = 11.84EE424 pKa = 3.98RR425 pKa = 11.84LWVEE429 pKa = 4.21FGLAMDD435 pKa = 4.52RR436 pKa = 11.84FAVCFEE442 pKa = 4.39DD443 pKa = 3.75VQGIPSPGEE452 pKa = 3.45PCLPFGEE459 pKa = 4.93GWSNLDD465 pKa = 3.36HH466 pKa = 6.53FRR468 pKa = 11.84EE469 pKa = 4.17YY470 pKa = 11.27LDD472 pKa = 4.13GLFKK476 pKa = 11.1VGLEE480 pKa = 4.05KK481 pKa = 10.57KK482 pKa = 9.21HH483 pKa = 5.37QNKK486 pKa = 6.99VTQNFPPWIATTNNYY501 pKa = 9.77IIPEE505 pKa = 4.33SVLEE509 pKa = 4.07RR510 pKa = 11.84AVVIPFKK517 pKa = 10.9RR518 pKa = 11.84MSFDD522 pKa = 3.14VQKK525 pKa = 10.91YY526 pKa = 9.4IEE528 pKa = 4.71EE529 pKa = 4.16FDD531 pKa = 3.25VDD533 pKa = 4.75LRR535 pKa = 11.84FLSSGPCLFLLWLLEE550 pKa = 4.36DD551 pKa = 4.13SVDD554 pKa = 5.48DD555 pKa = 5.45IDD557 pKa = 6.66DD558 pKa = 4.0EE559 pKa = 4.63LLDD562 pKa = 3.86RR563 pKa = 11.84AVVLSLAAKK572 pKa = 7.55MIWNAEE578 pKa = 3.63MRR580 pKa = 11.84DD581 pKa = 4.24FIKK584 pKa = 10.73APPPDD589 pKa = 3.97VLTAVYY595 pKa = 9.89VAEE598 pKa = 5.22DD599 pKa = 3.36GGIRR603 pKa = 11.84CTTGPEE609 pKa = 4.08KK610 pKa = 10.82EE611 pKa = 4.14NTPFTVSKK619 pKa = 9.63VWRR622 pKa = 11.84AIEE625 pKa = 4.08KK626 pKa = 10.6EE627 pKa = 4.1EE628 pKa = 4.03EE629 pKa = 3.92QLPEE633 pKa = 5.1DD634 pKa = 3.67IHH636 pKa = 5.79WTDD639 pKa = 3.65SEE641 pKa = 4.39SDD643 pKa = 3.72SVKK646 pKa = 10.83SVLATPLPFTPPDD659 pKa = 4.41PGLGSSDD666 pKa = 3.8EE667 pKa = 4.29EE668 pKa = 4.47EE669 pKa = 4.29EE670 pKa = 4.35VPPRR674 pKa = 11.84QIPKK678 pKa = 9.77KK679 pKa = 9.29RR680 pKa = 11.84KK681 pKa = 9.48CPFIDD686 pKa = 3.89EE687 pKa = 4.74AATEE691 pKa = 4.22CADD694 pKa = 4.95DD695 pKa = 4.08EE696 pKa = 4.63QSDD699 pKa = 3.82SSEE702 pKa = 4.25PYY704 pKa = 9.54PVPFEE709 pKa = 4.27KK710 pKa = 10.62KK711 pKa = 8.87KK712 pKa = 10.8GPRR715 pKa = 11.84VWVVVRR721 pKa = 11.84EE722 pKa = 4.19DD723 pKa = 3.51GAIGDD728 pKa = 4.5IEE730 pKa = 4.8NGPAPPPDD738 pKa = 3.69EE739 pKa = 4.18QSTSGFTDD747 pKa = 3.49ASGTTDD753 pKa = 3.87KK754 pKa = 11.22DD755 pKa = 3.88DD756 pKa = 5.08PDD758 pKa = 4.06VEE760 pKa = 4.61TSS762 pKa = 3.02
Molecular weight: 84.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0E3JE56|A0A0E3JE56_9POLY VP1 OS=Trematomus pennellii polyomavirus 1 OX=2170103 PE=3 SV=1
MM1 pKa = 7.58GIVIGILAGVSAGIEE16 pKa = 4.29VAVVTAAAAAAEE28 pKa = 4.72AIGAALATAADD39 pKa = 3.46IGLAAEE45 pKa = 4.31VGIAAAEE52 pKa = 4.06TAGEE56 pKa = 4.25LFGEE60 pKa = 4.43TAMAGDD66 pKa = 3.52IVGFGGLIEE75 pKa = 4.87GATEE79 pKa = 4.79PIAAAGMGVGGALQLAGIGATVASAVGTTVAALARR114 pKa = 11.84TRR116 pKa = 11.84AEE118 pKa = 3.69GDD120 pKa = 3.59VAHH123 pKa = 6.31VLVNAGTEE131 pKa = 3.73IQTAVTNIDD140 pKa = 3.27ASSTNARR147 pKa = 11.84VPIWLTAIMGTDD159 pKa = 3.17EE160 pKa = 4.21VLTNLGEE167 pKa = 4.71LEE169 pKa = 4.18NQFSNGRR176 pKa = 11.84LSYY179 pKa = 11.28DD180 pKa = 3.26FTLDD184 pKa = 2.99QATAIAAEE192 pKa = 4.31AEE194 pKa = 4.09QAGRR198 pKa = 11.84GGVHH202 pKa = 5.26YY203 pKa = 10.64HH204 pKa = 6.1PVEE207 pKa = 4.65LDD209 pKa = 3.11TALFEE214 pKa = 5.16KK215 pKa = 9.77MIKK218 pKa = 10.26HH219 pKa = 5.08STQTTFYY226 pKa = 10.39LPAKK230 pKa = 10.44NGMLLSKK237 pKa = 10.45RR238 pKa = 11.84DD239 pKa = 3.7TEE241 pKa = 4.3KK242 pKa = 11.16VINDD246 pKa = 2.52VHH248 pKa = 6.68ARR250 pKa = 11.84NTGGVDD256 pKa = 3.29AQVLGGKK263 pKa = 9.41NPFEE267 pKa = 4.71SWRR270 pKa = 11.84SMASAEE276 pKa = 3.88QDD278 pKa = 2.73KK279 pKa = 9.77WLKK282 pKa = 10.78DD283 pKa = 3.27IEE285 pKa = 4.29KK286 pKa = 10.54EE287 pKa = 3.75EE288 pKa = 4.29GKK290 pKa = 10.62AKK292 pKa = 9.88EE293 pKa = 3.85RR294 pKa = 11.84EE295 pKa = 3.91RR296 pKa = 11.84RR297 pKa = 11.84LEE299 pKa = 3.75NQLRR303 pKa = 11.84HH304 pKa = 5.2LTEE307 pKa = 3.5EE308 pKa = 3.9HH309 pKa = 6.02RR310 pKa = 11.84RR311 pKa = 11.84KK312 pKa = 10.25SRR314 pKa = 11.84AHH316 pKa = 6.42RR317 pKa = 11.84LLDD320 pKa = 3.51EE321 pKa = 4.89RR322 pKa = 11.84EE323 pKa = 4.12WEE325 pKa = 4.07NDD327 pKa = 2.79DD328 pKa = 4.75RR329 pKa = 11.84LAKK332 pKa = 10.51KK333 pKa = 10.44LEE335 pKa = 4.08KK336 pKa = 10.43DD337 pKa = 3.47LQKK340 pKa = 11.32VKK342 pKa = 10.88DD343 pKa = 4.2DD344 pKa = 3.59NDD346 pKa = 3.17KK347 pKa = 11.25RR348 pKa = 11.84EE349 pKa = 3.94QFLEE353 pKa = 4.15DD354 pKa = 2.96AVHH357 pKa = 6.3YY358 pKa = 10.39RR359 pKa = 11.84RR360 pKa = 11.84EE361 pKa = 4.12DD362 pKa = 3.48LFHH365 pKa = 6.46SRR367 pKa = 11.84SMAVFRR373 pKa = 11.84GAEE376 pKa = 4.11YY377 pKa = 10.14PEE379 pKa = 3.81EE380 pKa = 4.65WIGKK384 pKa = 4.72MTRR387 pKa = 11.84IRR389 pKa = 11.84DD390 pKa = 3.56EE391 pKa = 3.89WRR393 pKa = 11.84HH394 pKa = 4.59GVQAFEE400 pKa = 4.17VVLRR404 pKa = 11.84KK405 pKa = 9.83HH406 pKa = 6.44SEE408 pKa = 3.65HH409 pKa = 6.65PQRR412 pKa = 11.84GDD414 pKa = 3.1IPVSNAYY421 pKa = 9.91HH422 pKa = 6.88PALIRR427 pKa = 11.84YY428 pKa = 7.48RR429 pKa = 11.84RR430 pKa = 11.84RR431 pKa = 11.84HH432 pKa = 4.56WRR434 pKa = 11.84LKK436 pKa = 9.09KK437 pKa = 10.59AKK439 pKa = 9.27LVAVNTSDD447 pKa = 3.58GRR449 pKa = 11.84AHH451 pKa = 6.9EE452 pKa = 4.73VTLSWYY458 pKa = 8.93TPVGEE463 pKa = 4.49KK464 pKa = 9.96PVPAGLDD471 pKa = 3.06KK472 pKa = 11.18GFKK475 pKa = 9.56FVRR478 pKa = 11.84EE479 pKa = 4.11LTEE482 pKa = 3.94SQLSVLTDD490 pKa = 3.04YY491 pKa = 11.22LRR493 pKa = 11.84YY494 pKa = 8.91HH495 pKa = 5.46QAGFEE500 pKa = 4.09ISGGALNIPQRR511 pKa = 11.84LLVGALTLQTARR523 pKa = 11.84HH524 pKa = 5.25QSFKK528 pKa = 11.1APRR531 pKa = 11.84KK532 pKa = 9.54RR533 pKa = 11.84KK534 pKa = 9.61AIHH537 pKa = 6.35QDD539 pKa = 2.91APDD542 pKa = 3.27QGRR545 pKa = 11.84IRR547 pKa = 11.84SSKK550 pKa = 8.45RR551 pKa = 11.84RR552 pKa = 11.84KK553 pKa = 9.62RR554 pKa = 11.84RR555 pKa = 11.84TT556 pKa = 3.14
MM1 pKa = 7.58GIVIGILAGVSAGIEE16 pKa = 4.29VAVVTAAAAAAEE28 pKa = 4.72AIGAALATAADD39 pKa = 3.46IGLAAEE45 pKa = 4.31VGIAAAEE52 pKa = 4.06TAGEE56 pKa = 4.25LFGEE60 pKa = 4.43TAMAGDD66 pKa = 3.52IVGFGGLIEE75 pKa = 4.87GATEE79 pKa = 4.79PIAAAGMGVGGALQLAGIGATVASAVGTTVAALARR114 pKa = 11.84TRR116 pKa = 11.84AEE118 pKa = 3.69GDD120 pKa = 3.59VAHH123 pKa = 6.31VLVNAGTEE131 pKa = 3.73IQTAVTNIDD140 pKa = 3.27ASSTNARR147 pKa = 11.84VPIWLTAIMGTDD159 pKa = 3.17EE160 pKa = 4.21VLTNLGEE167 pKa = 4.71LEE169 pKa = 4.18NQFSNGRR176 pKa = 11.84LSYY179 pKa = 11.28DD180 pKa = 3.26FTLDD184 pKa = 2.99QATAIAAEE192 pKa = 4.31AEE194 pKa = 4.09QAGRR198 pKa = 11.84GGVHH202 pKa = 5.26YY203 pKa = 10.64HH204 pKa = 6.1PVEE207 pKa = 4.65LDD209 pKa = 3.11TALFEE214 pKa = 5.16KK215 pKa = 9.77MIKK218 pKa = 10.26HH219 pKa = 5.08STQTTFYY226 pKa = 10.39LPAKK230 pKa = 10.44NGMLLSKK237 pKa = 10.45RR238 pKa = 11.84DD239 pKa = 3.7TEE241 pKa = 4.3KK242 pKa = 11.16VINDD246 pKa = 2.52VHH248 pKa = 6.68ARR250 pKa = 11.84NTGGVDD256 pKa = 3.29AQVLGGKK263 pKa = 9.41NPFEE267 pKa = 4.71SWRR270 pKa = 11.84SMASAEE276 pKa = 3.88QDD278 pKa = 2.73KK279 pKa = 9.77WLKK282 pKa = 10.78DD283 pKa = 3.27IEE285 pKa = 4.29KK286 pKa = 10.54EE287 pKa = 3.75EE288 pKa = 4.29GKK290 pKa = 10.62AKK292 pKa = 9.88EE293 pKa = 3.85RR294 pKa = 11.84EE295 pKa = 3.91RR296 pKa = 11.84RR297 pKa = 11.84LEE299 pKa = 3.75NQLRR303 pKa = 11.84HH304 pKa = 5.2LTEE307 pKa = 3.5EE308 pKa = 3.9HH309 pKa = 6.02RR310 pKa = 11.84RR311 pKa = 11.84KK312 pKa = 10.25SRR314 pKa = 11.84AHH316 pKa = 6.42RR317 pKa = 11.84LLDD320 pKa = 3.51EE321 pKa = 4.89RR322 pKa = 11.84EE323 pKa = 4.12WEE325 pKa = 4.07NDD327 pKa = 2.79DD328 pKa = 4.75RR329 pKa = 11.84LAKK332 pKa = 10.51KK333 pKa = 10.44LEE335 pKa = 4.08KK336 pKa = 10.43DD337 pKa = 3.47LQKK340 pKa = 11.32VKK342 pKa = 10.88DD343 pKa = 4.2DD344 pKa = 3.59NDD346 pKa = 3.17KK347 pKa = 11.25RR348 pKa = 11.84EE349 pKa = 3.94QFLEE353 pKa = 4.15DD354 pKa = 2.96AVHH357 pKa = 6.3YY358 pKa = 10.39RR359 pKa = 11.84RR360 pKa = 11.84EE361 pKa = 4.12DD362 pKa = 3.48LFHH365 pKa = 6.46SRR367 pKa = 11.84SMAVFRR373 pKa = 11.84GAEE376 pKa = 4.11YY377 pKa = 10.14PEE379 pKa = 3.81EE380 pKa = 4.65WIGKK384 pKa = 4.72MTRR387 pKa = 11.84IRR389 pKa = 11.84DD390 pKa = 3.56EE391 pKa = 3.89WRR393 pKa = 11.84HH394 pKa = 4.59GVQAFEE400 pKa = 4.17VVLRR404 pKa = 11.84KK405 pKa = 9.83HH406 pKa = 6.44SEE408 pKa = 3.65HH409 pKa = 6.65PQRR412 pKa = 11.84GDD414 pKa = 3.1IPVSNAYY421 pKa = 9.91HH422 pKa = 6.88PALIRR427 pKa = 11.84YY428 pKa = 7.48RR429 pKa = 11.84RR430 pKa = 11.84RR431 pKa = 11.84HH432 pKa = 4.56WRR434 pKa = 11.84LKK436 pKa = 9.09KK437 pKa = 10.59AKK439 pKa = 9.27LVAVNTSDD447 pKa = 3.58GRR449 pKa = 11.84AHH451 pKa = 6.9EE452 pKa = 4.73VTLSWYY458 pKa = 8.93TPVGEE463 pKa = 4.49KK464 pKa = 9.96PVPAGLDD471 pKa = 3.06KK472 pKa = 11.18GFKK475 pKa = 9.56FVRR478 pKa = 11.84EE479 pKa = 4.11LTEE482 pKa = 3.94SQLSVLTDD490 pKa = 3.04YY491 pKa = 11.22LRR493 pKa = 11.84YY494 pKa = 8.91HH495 pKa = 5.46QAGFEE500 pKa = 4.09ISGGALNIPQRR511 pKa = 11.84LLVGALTLQTARR523 pKa = 11.84HH524 pKa = 5.25QSFKK528 pKa = 11.1APRR531 pKa = 11.84KK532 pKa = 9.54RR533 pKa = 11.84KK534 pKa = 9.61AIHH537 pKa = 6.35QDD539 pKa = 2.91APDD542 pKa = 3.27QGRR545 pKa = 11.84IRR547 pKa = 11.84SSKK550 pKa = 8.45RR551 pKa = 11.84RR552 pKa = 11.84KK553 pKa = 9.62RR554 pKa = 11.84RR555 pKa = 11.84TT556 pKa = 3.14
Molecular weight: 61.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1697 |
55 |
762 |
424.3 |
46.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.546 ± 1.562 | 1.591 ± 0.737 |
6.305 ± 0.789 | 6.953 ± 0.984 |
3.889 ± 0.534 | 7.484 ± 0.788 |
2.121 ± 0.548 | 3.712 ± 0.453 |
5.598 ± 0.354 | 8.014 ± 0.174 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.005 ± 0.745 | 3.771 ± 0.442 |
5.48 ± 1.169 | 4.007 ± 0.836 |
5.303 ± 1.263 | 6.6 ± 1.143 |
5.48 ± 0.265 | 6.895 ± 0.107 |
1.65 ± 0.154 | 2.593 ± 0.452 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
