
Lake Sarah-associated circular virus-12
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.4
Get precalculated fractions of proteins
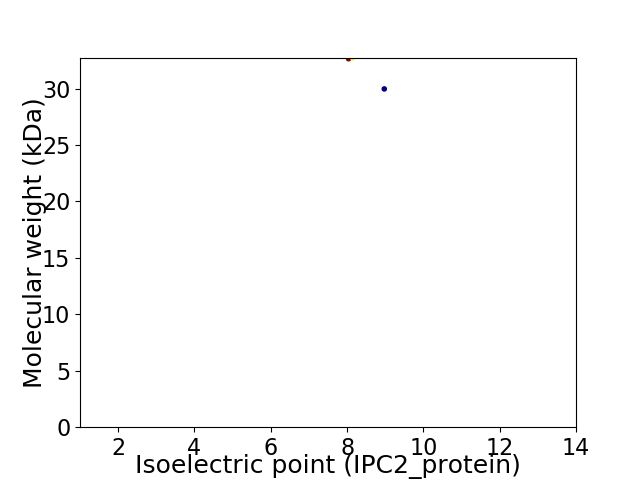
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
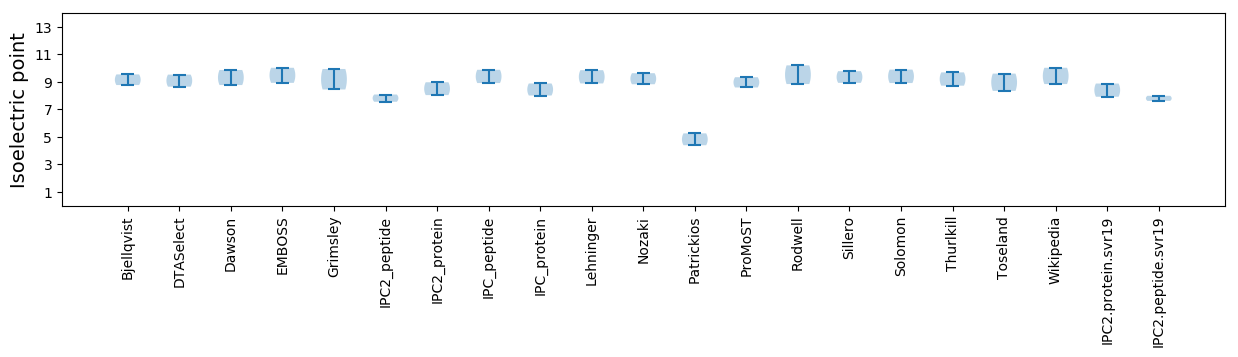
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQK4|A0A140AQK4_9VIRU Coat protein OS=Lake Sarah-associated circular virus-12 OX=1685738 PE=4 SV=1
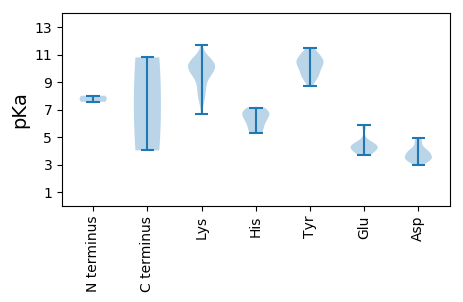
MM1 pKa = 8.01AYY3 pKa = 10.22NFCFTCNNPTDD14 pKa = 4.92AIEE17 pKa = 4.41TYY19 pKa = 9.46IQSITDD25 pKa = 3.15VRR27 pKa = 11.84FIGYY31 pKa = 8.68AHH33 pKa = 6.81EE34 pKa = 4.45VGEE37 pKa = 4.85LGTPHH42 pKa = 6.27LQGVVVLNKK51 pKa = 9.9KK52 pKa = 10.01SRR54 pKa = 11.84LTAVRR59 pKa = 11.84KK60 pKa = 9.53ILPKK64 pKa = 10.4CHH66 pKa = 6.5ISQMRR71 pKa = 11.84GSFAEE76 pKa = 3.94AWDD79 pKa = 4.7GYY81 pKa = 10.62ILTPHH86 pKa = 7.14AKK88 pKa = 9.6HH89 pKa = 6.11NKK91 pKa = 8.37PEE93 pKa = 4.26PEE95 pKa = 3.8VVYY98 pKa = 11.02AFGTPPGVRR107 pKa = 11.84GSNTRR112 pKa = 11.84DD113 pKa = 3.3VYY115 pKa = 11.02EE116 pKa = 3.95EE117 pKa = 3.83AMAIAKK123 pKa = 8.47TGTTEE128 pKa = 4.56GISAQMMLRR137 pKa = 11.84FGKK140 pKa = 10.18ALHH143 pKa = 6.39EE144 pKa = 4.46VAQKK148 pKa = 10.58HH149 pKa = 5.29SPKK152 pKa = 9.81PPHH155 pKa = 7.02IDD157 pKa = 3.51GDD159 pKa = 3.99LDD161 pKa = 3.77NYY163 pKa = 9.53WIYY166 pKa = 11.27GPTGSGKK173 pKa = 10.69SKK175 pKa = 9.47TVYY178 pKa = 10.49DD179 pKa = 3.35KK180 pKa = 10.76WPRR183 pKa = 11.84DD184 pKa = 3.26TLYY187 pKa = 10.59IKK189 pKa = 10.4LHH191 pKa = 5.32NKK193 pKa = 6.68WWEE196 pKa = 4.0NYY198 pKa = 9.85DD199 pKa = 3.62GEE201 pKa = 4.42EE202 pKa = 3.69TVYY205 pKa = 10.6IEE207 pKa = 5.87DD208 pKa = 4.71LGNDD212 pKa = 3.72SKK214 pKa = 11.7LLTHH218 pKa = 6.87IKK220 pKa = 9.72HH221 pKa = 5.44WADD224 pKa = 3.54RR225 pKa = 11.84YY226 pKa = 10.57AFRR229 pKa = 11.84AEE231 pKa = 4.14VKK233 pKa = 10.27FGSILIRR240 pKa = 11.84PKK242 pKa = 10.6RR243 pKa = 11.84LVVTSNHH250 pKa = 6.21TIRR253 pKa = 11.84DD254 pKa = 4.0SFPTASVADD263 pKa = 3.86VEE265 pKa = 4.23ALEE268 pKa = 4.26RR269 pKa = 11.84RR270 pKa = 11.84FKK272 pKa = 10.39IQYY275 pKa = 9.44IGPQIPAFTSVTKK288 pKa = 10.79
MM1 pKa = 8.01AYY3 pKa = 10.22NFCFTCNNPTDD14 pKa = 4.92AIEE17 pKa = 4.41TYY19 pKa = 9.46IQSITDD25 pKa = 3.15VRR27 pKa = 11.84FIGYY31 pKa = 8.68AHH33 pKa = 6.81EE34 pKa = 4.45VGEE37 pKa = 4.85LGTPHH42 pKa = 6.27LQGVVVLNKK51 pKa = 9.9KK52 pKa = 10.01SRR54 pKa = 11.84LTAVRR59 pKa = 11.84KK60 pKa = 9.53ILPKK64 pKa = 10.4CHH66 pKa = 6.5ISQMRR71 pKa = 11.84GSFAEE76 pKa = 3.94AWDD79 pKa = 4.7GYY81 pKa = 10.62ILTPHH86 pKa = 7.14AKK88 pKa = 9.6HH89 pKa = 6.11NKK91 pKa = 8.37PEE93 pKa = 4.26PEE95 pKa = 3.8VVYY98 pKa = 11.02AFGTPPGVRR107 pKa = 11.84GSNTRR112 pKa = 11.84DD113 pKa = 3.3VYY115 pKa = 11.02EE116 pKa = 3.95EE117 pKa = 3.83AMAIAKK123 pKa = 8.47TGTTEE128 pKa = 4.56GISAQMMLRR137 pKa = 11.84FGKK140 pKa = 10.18ALHH143 pKa = 6.39EE144 pKa = 4.46VAQKK148 pKa = 10.58HH149 pKa = 5.29SPKK152 pKa = 9.81PPHH155 pKa = 7.02IDD157 pKa = 3.51GDD159 pKa = 3.99LDD161 pKa = 3.77NYY163 pKa = 9.53WIYY166 pKa = 11.27GPTGSGKK173 pKa = 10.69SKK175 pKa = 9.47TVYY178 pKa = 10.49DD179 pKa = 3.35KK180 pKa = 10.76WPRR183 pKa = 11.84DD184 pKa = 3.26TLYY187 pKa = 10.59IKK189 pKa = 10.4LHH191 pKa = 5.32NKK193 pKa = 6.68WWEE196 pKa = 4.0NYY198 pKa = 9.85DD199 pKa = 3.62GEE201 pKa = 4.42EE202 pKa = 3.69TVYY205 pKa = 10.6IEE207 pKa = 5.87DD208 pKa = 4.71LGNDD212 pKa = 3.72SKK214 pKa = 11.7LLTHH218 pKa = 6.87IKK220 pKa = 9.72HH221 pKa = 5.44WADD224 pKa = 3.54RR225 pKa = 11.84YY226 pKa = 10.57AFRR229 pKa = 11.84AEE231 pKa = 4.14VKK233 pKa = 10.27FGSILIRR240 pKa = 11.84PKK242 pKa = 10.6RR243 pKa = 11.84LVVTSNHH250 pKa = 6.21TIRR253 pKa = 11.84DD254 pKa = 4.0SFPTASVADD263 pKa = 3.86VEE265 pKa = 4.23ALEE268 pKa = 4.26RR269 pKa = 11.84RR270 pKa = 11.84FKK272 pKa = 10.39IQYY275 pKa = 9.44IGPQIPAFTSVTKK288 pKa = 10.79
Molecular weight: 32.64 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQK4|A0A140AQK4_9VIRU Coat protein OS=Lake Sarah-associated circular virus-12 OX=1685738 PE=4 SV=1
MM1 pKa = 7.56PAVKK5 pKa = 8.92KK6 pKa = 8.07TFRR9 pKa = 11.84KK10 pKa = 9.68GKK12 pKa = 7.8SAKK15 pKa = 8.82RR16 pKa = 11.84TSNKK20 pKa = 7.77STKK23 pKa = 7.68TKK25 pKa = 10.17KK26 pKa = 9.99PKK28 pKa = 9.64ASKK31 pKa = 9.89VFKK34 pKa = 10.59KK35 pKa = 10.38KK36 pKa = 9.4VEE38 pKa = 4.01RR39 pKa = 11.84VIASQAEE46 pKa = 4.4VKK48 pKa = 10.55HH49 pKa = 5.84ISTWGAPAYY58 pKa = 9.96YY59 pKa = 10.33NSAIAGAGDD68 pKa = 3.94INPILPPMFQGTKK81 pKa = 9.2DD82 pKa = 3.32RR83 pKa = 11.84EE84 pKa = 4.37RR85 pKa = 11.84IGSKK89 pKa = 10.23VNFRR93 pKa = 11.84SFRR96 pKa = 11.84LNICISQAPYY106 pKa = 11.44ALLTDD111 pKa = 4.25LDD113 pKa = 4.06RR114 pKa = 11.84PVLVRR119 pKa = 11.84LLVLTYY125 pKa = 10.64KK126 pKa = 10.34RR127 pKa = 11.84GKK129 pKa = 8.5YY130 pKa = 9.23QPDD133 pKa = 3.49FTTSRR138 pKa = 11.84IVTDD142 pKa = 3.55VNEE145 pKa = 4.42SLLHH149 pKa = 5.73QDD151 pKa = 3.65ASAPAQPYY159 pKa = 10.03DD160 pKa = 3.95GSILHH165 pKa = 6.7HH166 pKa = 6.77LFPVNNDD173 pKa = 2.95LFTVIKK179 pKa = 10.24DD180 pKa = 3.13ISFTILPNEE189 pKa = 4.36LNDD192 pKa = 3.39NTTNSQTTALGKK204 pKa = 9.96NFRR207 pKa = 11.84TFSVNVPGPSHH218 pKa = 6.86LVYY221 pKa = 11.02GEE223 pKa = 5.7DD224 pKa = 3.46INQYY228 pKa = 9.56PNNFAPFFSVGYY240 pKa = 9.1CYY242 pKa = 10.09PDD244 pKa = 4.11GDD246 pKa = 4.57LADD249 pKa = 3.83VLNTGLRR256 pKa = 11.84VTWEE260 pKa = 3.93SRR262 pKa = 11.84LSYY265 pKa = 10.38TDD267 pKa = 3.32EE268 pKa = 4.03
MM1 pKa = 7.56PAVKK5 pKa = 8.92KK6 pKa = 8.07TFRR9 pKa = 11.84KK10 pKa = 9.68GKK12 pKa = 7.8SAKK15 pKa = 8.82RR16 pKa = 11.84TSNKK20 pKa = 7.77STKK23 pKa = 7.68TKK25 pKa = 10.17KK26 pKa = 9.99PKK28 pKa = 9.64ASKK31 pKa = 9.89VFKK34 pKa = 10.59KK35 pKa = 10.38KK36 pKa = 9.4VEE38 pKa = 4.01RR39 pKa = 11.84VIASQAEE46 pKa = 4.4VKK48 pKa = 10.55HH49 pKa = 5.84ISTWGAPAYY58 pKa = 9.96YY59 pKa = 10.33NSAIAGAGDD68 pKa = 3.94INPILPPMFQGTKK81 pKa = 9.2DD82 pKa = 3.32RR83 pKa = 11.84EE84 pKa = 4.37RR85 pKa = 11.84IGSKK89 pKa = 10.23VNFRR93 pKa = 11.84SFRR96 pKa = 11.84LNICISQAPYY106 pKa = 11.44ALLTDD111 pKa = 4.25LDD113 pKa = 4.06RR114 pKa = 11.84PVLVRR119 pKa = 11.84LLVLTYY125 pKa = 10.64KK126 pKa = 10.34RR127 pKa = 11.84GKK129 pKa = 8.5YY130 pKa = 9.23QPDD133 pKa = 3.49FTTSRR138 pKa = 11.84IVTDD142 pKa = 3.55VNEE145 pKa = 4.42SLLHH149 pKa = 5.73QDD151 pKa = 3.65ASAPAQPYY159 pKa = 10.03DD160 pKa = 3.95GSILHH165 pKa = 6.7HH166 pKa = 6.77LFPVNNDD173 pKa = 2.95LFTVIKK179 pKa = 10.24DD180 pKa = 3.13ISFTILPNEE189 pKa = 4.36LNDD192 pKa = 3.39NTTNSQTTALGKK204 pKa = 9.96NFRR207 pKa = 11.84TFSVNVPGPSHH218 pKa = 6.86LVYY221 pKa = 11.02GEE223 pKa = 5.7DD224 pKa = 3.46INQYY228 pKa = 9.56PNNFAPFFSVGYY240 pKa = 9.1CYY242 pKa = 10.09PDD244 pKa = 4.11GDD246 pKa = 4.57LADD249 pKa = 3.83VLNTGLRR256 pKa = 11.84VTWEE260 pKa = 3.93SRR262 pKa = 11.84LSYY265 pKa = 10.38TDD267 pKa = 3.32EE268 pKa = 4.03
Molecular weight: 29.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
556 |
268 |
288 |
278.0 |
31.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.014 ± 0.209 | 0.899 ± 0.107 |
5.576 ± 0.277 | 4.496 ± 1.062 |
4.496 ± 0.511 | 6.115 ± 0.626 |
3.058 ± 0.837 | 6.115 ± 0.626 |
7.554 ± 0.198 | 7.014 ± 0.839 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.259 ± 0.36 | 5.216 ± 1.054 |
6.295 ± 0.296 | 2.698 ± 0.202 |
5.216 ± 0.006 | 6.475 ± 0.956 |
7.734 ± 0.072 | 6.835 ± 0.179 |
1.439 ± 0.486 | 4.496 ± 0.275 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
