
McMurdo Ice Shelf pond-associated circular DNA virus-8
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.55
Get precalculated fractions of proteins
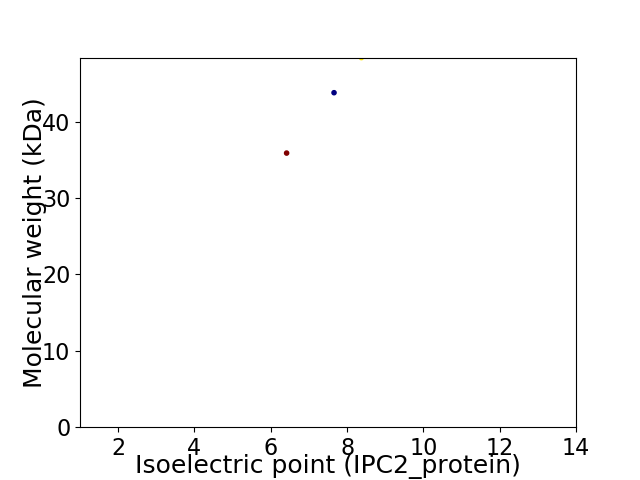
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A075LZ83|A0A075LZ83_9VIRU Uncharacterized protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-8 OX=1521392 PE=4 SV=1
MM1 pKa = 7.5CGRR4 pKa = 11.84WGFRR8 pKa = 11.84DD9 pKa = 3.46WDD11 pKa = 3.71PVEE14 pKa = 4.03ARR16 pKa = 11.84LYY18 pKa = 10.3QPTVQKK24 pKa = 10.97VLEE27 pKa = 5.01DD28 pKa = 4.4YY29 pKa = 11.35GDD31 pKa = 3.47DD32 pKa = 3.82WIFQLEE38 pKa = 4.15MGEE41 pKa = 4.39SGFVHH46 pKa = 5.66YY47 pKa = 10.22QIYY50 pKa = 9.13WKK52 pKa = 8.98LTKK55 pKa = 9.51RR56 pKa = 11.84DD57 pKa = 3.67RR58 pKa = 11.84PKK60 pKa = 10.39TLAIALNEE68 pKa = 3.92RR69 pKa = 11.84LPGIEE74 pKa = 4.88LSAASTGGVLALKK87 pKa = 9.61RR88 pKa = 11.84YY89 pKa = 9.12CMKK92 pKa = 10.84DD93 pKa = 3.25DD94 pKa = 3.58TRR96 pKa = 11.84LAGPWSKK103 pKa = 11.07KK104 pKa = 9.59KK105 pKa = 10.15IYY107 pKa = 10.08RR108 pKa = 11.84GEE110 pKa = 4.17DD111 pKa = 3.48LPVKK115 pKa = 9.45MLPWQQKK122 pKa = 9.16LADD125 pKa = 3.97YY126 pKa = 10.81LKK128 pKa = 9.17TEE130 pKa = 4.0AGKK133 pKa = 10.7RR134 pKa = 11.84EE135 pKa = 4.71IIWLMDD141 pKa = 4.06PLGKK145 pKa = 9.23MGKK148 pKa = 8.12TIFGKK153 pKa = 9.99MMAYY157 pKa = 8.86WRR159 pKa = 11.84GALYY163 pKa = 8.06LTWGDD168 pKa = 3.68TKK170 pKa = 11.54DD171 pKa = 4.11VMNLVAKK178 pKa = 10.16SDD180 pKa = 3.73KK181 pKa = 10.69NNVYY185 pKa = 10.1IFNLSRR191 pKa = 11.84TKK193 pKa = 10.48PKK195 pKa = 10.25MMSSDD200 pKa = 3.91DD201 pKa = 4.48LYY203 pKa = 11.17TALEE207 pKa = 4.1QIKK210 pKa = 10.65DD211 pKa = 3.46GAFINTKK218 pKa = 10.38YY219 pKa = 8.27EE220 pKa = 4.03TSQVLMKK227 pKa = 9.75PPHH230 pKa = 5.65VVVMANCMPEE240 pKa = 3.85MAKK243 pKa = 10.39LSKK246 pKa = 10.66DD247 pKa = 2.88RR248 pKa = 11.84WNVIVMKK255 pKa = 10.62FSDD258 pKa = 3.94RR259 pKa = 11.84PPVEE263 pKa = 3.75EE264 pKa = 3.77EE265 pKa = 3.96TRR267 pKa = 11.84FTMEE271 pKa = 4.4DD272 pKa = 3.27PVKK275 pKa = 9.96EE276 pKa = 4.06RR277 pKa = 11.84EE278 pKa = 4.38GGGSSSQEE286 pKa = 3.87TVVLSQEE293 pKa = 3.97EE294 pKa = 4.61HH295 pKa = 6.23EE296 pKa = 4.63VMGDD300 pKa = 3.39QFPDD304 pKa = 3.25SSAPTGASVV313 pKa = 3.48
MM1 pKa = 7.5CGRR4 pKa = 11.84WGFRR8 pKa = 11.84DD9 pKa = 3.46WDD11 pKa = 3.71PVEE14 pKa = 4.03ARR16 pKa = 11.84LYY18 pKa = 10.3QPTVQKK24 pKa = 10.97VLEE27 pKa = 5.01DD28 pKa = 4.4YY29 pKa = 11.35GDD31 pKa = 3.47DD32 pKa = 3.82WIFQLEE38 pKa = 4.15MGEE41 pKa = 4.39SGFVHH46 pKa = 5.66YY47 pKa = 10.22QIYY50 pKa = 9.13WKK52 pKa = 8.98LTKK55 pKa = 9.51RR56 pKa = 11.84DD57 pKa = 3.67RR58 pKa = 11.84PKK60 pKa = 10.39TLAIALNEE68 pKa = 3.92RR69 pKa = 11.84LPGIEE74 pKa = 4.88LSAASTGGVLALKK87 pKa = 9.61RR88 pKa = 11.84YY89 pKa = 9.12CMKK92 pKa = 10.84DD93 pKa = 3.25DD94 pKa = 3.58TRR96 pKa = 11.84LAGPWSKK103 pKa = 11.07KK104 pKa = 9.59KK105 pKa = 10.15IYY107 pKa = 10.08RR108 pKa = 11.84GEE110 pKa = 4.17DD111 pKa = 3.48LPVKK115 pKa = 9.45MLPWQQKK122 pKa = 9.16LADD125 pKa = 3.97YY126 pKa = 10.81LKK128 pKa = 9.17TEE130 pKa = 4.0AGKK133 pKa = 10.7RR134 pKa = 11.84EE135 pKa = 4.71IIWLMDD141 pKa = 4.06PLGKK145 pKa = 9.23MGKK148 pKa = 8.12TIFGKK153 pKa = 9.99MMAYY157 pKa = 8.86WRR159 pKa = 11.84GALYY163 pKa = 8.06LTWGDD168 pKa = 3.68TKK170 pKa = 11.54DD171 pKa = 4.11VMNLVAKK178 pKa = 10.16SDD180 pKa = 3.73KK181 pKa = 10.69NNVYY185 pKa = 10.1IFNLSRR191 pKa = 11.84TKK193 pKa = 10.48PKK195 pKa = 10.25MMSSDD200 pKa = 3.91DD201 pKa = 4.48LYY203 pKa = 11.17TALEE207 pKa = 4.1QIKK210 pKa = 10.65DD211 pKa = 3.46GAFINTKK218 pKa = 10.38YY219 pKa = 8.27EE220 pKa = 4.03TSQVLMKK227 pKa = 9.75PPHH230 pKa = 5.65VVVMANCMPEE240 pKa = 3.85MAKK243 pKa = 10.39LSKK246 pKa = 10.66DD247 pKa = 2.88RR248 pKa = 11.84WNVIVMKK255 pKa = 10.62FSDD258 pKa = 3.94RR259 pKa = 11.84PPVEE263 pKa = 3.75EE264 pKa = 3.77EE265 pKa = 3.96TRR267 pKa = 11.84FTMEE271 pKa = 4.4DD272 pKa = 3.27PVKK275 pKa = 9.96EE276 pKa = 4.06RR277 pKa = 11.84EE278 pKa = 4.38GGGSSSQEE286 pKa = 3.87TVVLSQEE293 pKa = 3.97EE294 pKa = 4.61HH295 pKa = 6.23EE296 pKa = 4.63VMGDD300 pKa = 3.39QFPDD304 pKa = 3.25SSAPTGASVV313 pKa = 3.48
Molecular weight: 35.88 kDa
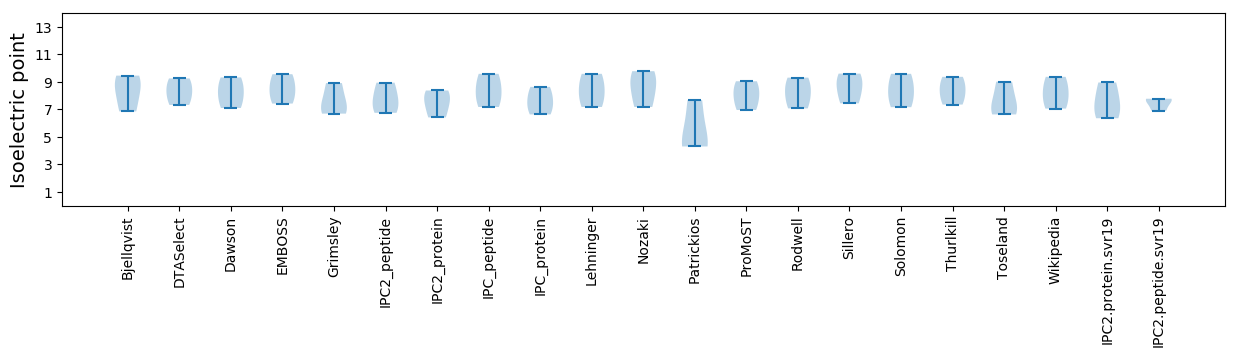
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A075LZ83|A0A075LZ83_9VIRU Uncharacterized protein OS=McMurdo Ice Shelf pond-associated circular DNA virus-8 OX=1521392 PE=4 SV=1
MM1 pKa = 7.23GRR3 pKa = 11.84DD4 pKa = 3.51VTPTSWTTCLEE15 pKa = 3.87GEE17 pKa = 4.65MYY19 pKa = 10.73GSHH22 pKa = 5.82TRR24 pKa = 11.84NHH26 pKa = 7.16DD27 pKa = 3.64SQLLDD32 pKa = 3.15SSEE35 pKa = 4.04IVSGEE40 pKa = 3.78RR41 pKa = 11.84IDD43 pKa = 5.33RR44 pKa = 11.84VGSDD48 pKa = 3.38LRR50 pKa = 11.84LARR53 pKa = 11.84ALEE56 pKa = 4.33GVGLARR62 pKa = 11.84KK63 pKa = 9.78SSALSLLGNSGTLGLHH79 pKa = 6.32GDD81 pKa = 4.11DD82 pKa = 3.39VGVLNGVPRR91 pKa = 11.84NLNRR95 pKa = 11.84EE96 pKa = 4.08LQTDD100 pKa = 4.35GEE102 pKa = 4.49LGLNGGGAGMTPPDD116 pKa = 3.64PRR118 pKa = 11.84AVEE121 pKa = 4.11RR122 pKa = 11.84VLSGRR127 pKa = 11.84RR128 pKa = 11.84HH129 pKa = 5.8HH130 pKa = 6.78NGGVDD135 pKa = 3.23RR136 pKa = 11.84RR137 pKa = 11.84KK138 pKa = 8.38TRR140 pKa = 11.84VLDD143 pKa = 3.45EE144 pKa = 4.19VLAVLIVDD152 pKa = 3.14QHH154 pKa = 7.28SGVRR158 pKa = 11.84FTADD162 pKa = 3.35DD163 pKa = 4.53TITLANRR170 pKa = 11.84HH171 pKa = 5.76NSLVSPTLDD180 pKa = 3.08VRR182 pKa = 11.84EE183 pKa = 3.87VDD185 pKa = 3.52GRR187 pKa = 11.84EE188 pKa = 3.9TRR190 pKa = 11.84SGSIRR195 pKa = 11.84VRR197 pKa = 11.84VAGMRR202 pKa = 11.84AAEE205 pKa = 4.3DD206 pKa = 3.21QTTVVRR212 pKa = 11.84LRR214 pKa = 11.84GDD216 pKa = 3.3SSIVRR221 pKa = 11.84VHH223 pKa = 6.38HH224 pKa = 5.87NRR226 pKa = 11.84VGEE229 pKa = 4.08RR230 pKa = 11.84NHH232 pKa = 6.24AHH234 pKa = 6.97LKK236 pKa = 9.91RR237 pKa = 11.84VNSHH241 pKa = 5.85PLEE244 pKa = 4.32AVVEE248 pKa = 4.19VLALFRR254 pKa = 11.84EE255 pKa = 4.61VEE257 pKa = 3.97EE258 pKa = 5.04GGAEE262 pKa = 3.99RR263 pKa = 11.84TDD265 pKa = 3.59TGQGTAQEE273 pKa = 4.24QLEE276 pKa = 4.51SLSTTSSGVGHH287 pKa = 6.89GSSEE291 pKa = 3.7EE292 pKa = 3.73RR293 pKa = 11.84RR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84SAQGGRR302 pKa = 11.84VGVVHH307 pKa = 6.79LVGAGDD313 pKa = 4.29LATVQPGVRR322 pKa = 11.84HH323 pKa = 6.57RR324 pKa = 11.84DD325 pKa = 3.33RR326 pKa = 11.84PHH328 pKa = 6.37EE329 pKa = 4.22VGGTPLRR336 pKa = 11.84GTLTPTSLSTAVLTGRR352 pKa = 11.84DD353 pKa = 3.96GACRR357 pKa = 11.84ALACPFLWNDD367 pKa = 3.27LEE369 pKa = 4.34IVVVGQIGAFHH380 pKa = 7.01RR381 pKa = 11.84RR382 pKa = 11.84QGRR385 pKa = 11.84QGCLRR390 pKa = 11.84ALLRR394 pKa = 11.84VLEE397 pKa = 4.33LVVPRR402 pKa = 11.84DD403 pKa = 3.34GSSWRR408 pKa = 11.84RR409 pKa = 11.84TDD411 pKa = 3.61RR412 pKa = 11.84RR413 pKa = 11.84MGTLKK418 pKa = 10.44RR419 pKa = 11.84RR420 pKa = 11.84LCEE423 pKa = 4.35GGNGPHH429 pKa = 5.6TTSTRR434 pKa = 11.84IWFMDD439 pKa = 3.8LFQNTSDD446 pKa = 3.35
MM1 pKa = 7.23GRR3 pKa = 11.84DD4 pKa = 3.51VTPTSWTTCLEE15 pKa = 3.87GEE17 pKa = 4.65MYY19 pKa = 10.73GSHH22 pKa = 5.82TRR24 pKa = 11.84NHH26 pKa = 7.16DD27 pKa = 3.64SQLLDD32 pKa = 3.15SSEE35 pKa = 4.04IVSGEE40 pKa = 3.78RR41 pKa = 11.84IDD43 pKa = 5.33RR44 pKa = 11.84VGSDD48 pKa = 3.38LRR50 pKa = 11.84LARR53 pKa = 11.84ALEE56 pKa = 4.33GVGLARR62 pKa = 11.84KK63 pKa = 9.78SSALSLLGNSGTLGLHH79 pKa = 6.32GDD81 pKa = 4.11DD82 pKa = 3.39VGVLNGVPRR91 pKa = 11.84NLNRR95 pKa = 11.84EE96 pKa = 4.08LQTDD100 pKa = 4.35GEE102 pKa = 4.49LGLNGGGAGMTPPDD116 pKa = 3.64PRR118 pKa = 11.84AVEE121 pKa = 4.11RR122 pKa = 11.84VLSGRR127 pKa = 11.84RR128 pKa = 11.84HH129 pKa = 5.8HH130 pKa = 6.78NGGVDD135 pKa = 3.23RR136 pKa = 11.84RR137 pKa = 11.84KK138 pKa = 8.38TRR140 pKa = 11.84VLDD143 pKa = 3.45EE144 pKa = 4.19VLAVLIVDD152 pKa = 3.14QHH154 pKa = 7.28SGVRR158 pKa = 11.84FTADD162 pKa = 3.35DD163 pKa = 4.53TITLANRR170 pKa = 11.84HH171 pKa = 5.76NSLVSPTLDD180 pKa = 3.08VRR182 pKa = 11.84EE183 pKa = 3.87VDD185 pKa = 3.52GRR187 pKa = 11.84EE188 pKa = 3.9TRR190 pKa = 11.84SGSIRR195 pKa = 11.84VRR197 pKa = 11.84VAGMRR202 pKa = 11.84AAEE205 pKa = 4.3DD206 pKa = 3.21QTTVVRR212 pKa = 11.84LRR214 pKa = 11.84GDD216 pKa = 3.3SSIVRR221 pKa = 11.84VHH223 pKa = 6.38HH224 pKa = 5.87NRR226 pKa = 11.84VGEE229 pKa = 4.08RR230 pKa = 11.84NHH232 pKa = 6.24AHH234 pKa = 6.97LKK236 pKa = 9.91RR237 pKa = 11.84VNSHH241 pKa = 5.85PLEE244 pKa = 4.32AVVEE248 pKa = 4.19VLALFRR254 pKa = 11.84EE255 pKa = 4.61VEE257 pKa = 3.97EE258 pKa = 5.04GGAEE262 pKa = 3.99RR263 pKa = 11.84TDD265 pKa = 3.59TGQGTAQEE273 pKa = 4.24QLEE276 pKa = 4.51SLSTTSSGVGHH287 pKa = 6.89GSSEE291 pKa = 3.7EE292 pKa = 3.73RR293 pKa = 11.84RR294 pKa = 11.84RR295 pKa = 11.84RR296 pKa = 11.84SAQGGRR302 pKa = 11.84VGVVHH307 pKa = 6.79LVGAGDD313 pKa = 4.29LATVQPGVRR322 pKa = 11.84HH323 pKa = 6.57RR324 pKa = 11.84DD325 pKa = 3.33RR326 pKa = 11.84PHH328 pKa = 6.37EE329 pKa = 4.22VGGTPLRR336 pKa = 11.84GTLTPTSLSTAVLTGRR352 pKa = 11.84DD353 pKa = 3.96GACRR357 pKa = 11.84ALACPFLWNDD367 pKa = 3.27LEE369 pKa = 4.34IVVVGQIGAFHH380 pKa = 7.01RR381 pKa = 11.84RR382 pKa = 11.84QGRR385 pKa = 11.84QGCLRR390 pKa = 11.84ALLRR394 pKa = 11.84VLEE397 pKa = 4.33LVVPRR402 pKa = 11.84DD403 pKa = 3.34GSSWRR408 pKa = 11.84RR409 pKa = 11.84TDD411 pKa = 3.61RR412 pKa = 11.84RR413 pKa = 11.84MGTLKK418 pKa = 10.44RR419 pKa = 11.84RR420 pKa = 11.84LCEE423 pKa = 4.35GGNGPHH429 pKa = 5.6TTSTRR434 pKa = 11.84IWFMDD439 pKa = 3.8LFQNTSDD446 pKa = 3.35
Molecular weight: 48.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1167 |
313 |
446 |
389.0 |
42.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.141 ± 1.806 | 1.285 ± 0.178 |
5.741 ± 0.567 | 5.484 ± 0.754 |
2.656 ± 0.651 | 8.997 ± 1.525 |
2.485 ± 0.721 | 3.342 ± 0.625 |
4.199 ± 1.732 | 8.483 ± 1.164 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.742 ± 0.966 | 3.428 ± 0.334 |
4.97 ± 0.889 | 3.599 ± 0.375 |
7.883 ± 2.136 | 7.198 ± 0.481 |
7.369 ± 0.647 | 8.226 ± 0.85 |
1.628 ± 0.503 | 2.142 ± 0.88 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
