
Bat cyclovirus GF-4c
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Cyclovirus; Bat associated cyclovirus 1
Average proteome isoelectric point is 8.54
Get precalculated fractions of proteins
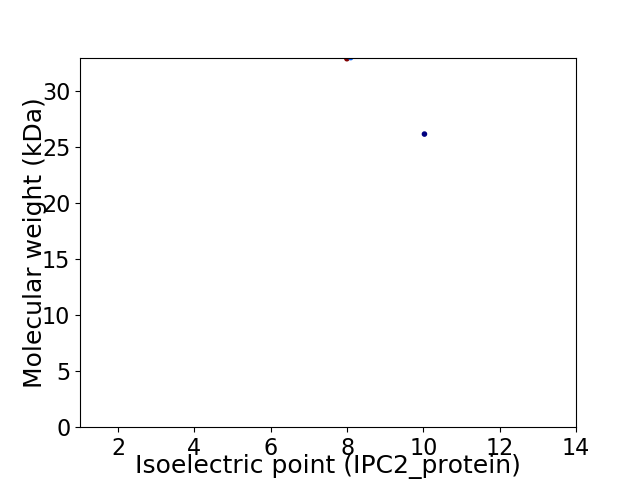
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D9IZ81|D9IZ81_9CIRC Putative Cap OS=Bat cyclovirus GF-4c OX=795381 PE=4 SV=1
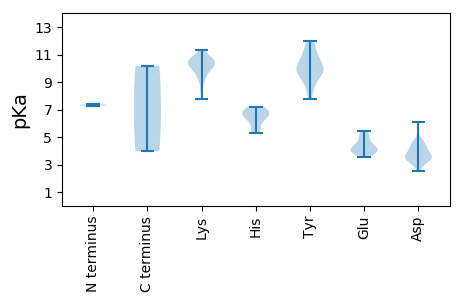
MM1 pKa = 7.37EE2 pKa = 5.26NKK4 pKa = 9.6TIRR7 pKa = 11.84RR8 pKa = 11.84FIFTWNNYY16 pKa = 8.38TDD18 pKa = 4.57DD19 pKa = 6.08DD20 pKa = 4.61YY21 pKa = 11.99DD22 pKa = 3.72KK23 pKa = 10.66TCAFIKK29 pKa = 10.29GNCKK33 pKa = 10.03YY34 pKa = 10.73GIVGKK39 pKa = 9.26EE40 pKa = 3.74HH41 pKa = 7.2APTTGVPHH49 pKa = 6.69LQGFCNLSKK58 pKa = 10.66PMRR61 pKa = 11.84FSAIKK66 pKa = 9.89KK67 pKa = 8.67CLSNSIHH74 pKa = 6.31IEE76 pKa = 3.76KK77 pKa = 10.55AIGSDD82 pKa = 3.42EE83 pKa = 4.04QNQKK87 pKa = 10.03YY88 pKa = 9.71CSKK91 pKa = 10.96AGDD94 pKa = 3.8FFEE97 pKa = 5.43EE98 pKa = 4.57GLPYY102 pKa = 9.93TQGKK106 pKa = 7.77RR107 pKa = 11.84TDD109 pKa = 3.5LEE111 pKa = 4.07AVVRR115 pKa = 11.84DD116 pKa = 3.94MEE118 pKa = 4.35QGTVRR123 pKa = 11.84LEE125 pKa = 4.59DD126 pKa = 3.15IAAKK130 pKa = 10.13HH131 pKa = 5.52PCAFIKK137 pKa = 8.34YY138 pKa = 9.62HH139 pKa = 6.87RR140 pKa = 11.84GIDD143 pKa = 3.5QLLKK147 pKa = 10.34LIHH150 pKa = 6.8PIKK153 pKa = 10.54PRR155 pKa = 11.84DD156 pKa = 3.56FQTYY160 pKa = 7.78VYY162 pKa = 9.12YY163 pKa = 10.77YY164 pKa = 8.96WGEE167 pKa = 4.08PGTGKK172 pKa = 10.02SRR174 pKa = 11.84RR175 pKa = 11.84AKK177 pKa = 10.62SEE179 pKa = 3.58ADD181 pKa = 3.28AVNSASIYY189 pKa = 8.73YY190 pKa = 9.87KK191 pKa = 10.51PRR193 pKa = 11.84GLWWDD198 pKa = 4.4GYY200 pKa = 9.51RR201 pKa = 11.84QQEE204 pKa = 4.04NVIIDD209 pKa = 4.38DD210 pKa = 4.37FYY212 pKa = 11.73GWIKK216 pKa = 10.75YY217 pKa = 10.28DD218 pKa = 4.1EE219 pKa = 4.44LLKK222 pKa = 10.71ICDD225 pKa = 3.53RR226 pKa = 11.84YY227 pKa = 9.88PYY229 pKa = 10.1KK230 pKa = 10.94VQIKK234 pKa = 10.43GGFEE238 pKa = 3.92EE239 pKa = 4.93FTSKK243 pKa = 10.88RR244 pKa = 11.84IWITSNVDD252 pKa = 2.54IYY254 pKa = 11.62SLYY257 pKa = 10.75KK258 pKa = 10.27FEE260 pKa = 5.17GYY262 pKa = 9.61NAEE265 pKa = 3.93AFKK268 pKa = 10.98RR269 pKa = 11.84RR270 pKa = 11.84ITCMVEE276 pKa = 3.78MNSNKK281 pKa = 10.17
MM1 pKa = 7.37EE2 pKa = 5.26NKK4 pKa = 9.6TIRR7 pKa = 11.84RR8 pKa = 11.84FIFTWNNYY16 pKa = 8.38TDD18 pKa = 4.57DD19 pKa = 6.08DD20 pKa = 4.61YY21 pKa = 11.99DD22 pKa = 3.72KK23 pKa = 10.66TCAFIKK29 pKa = 10.29GNCKK33 pKa = 10.03YY34 pKa = 10.73GIVGKK39 pKa = 9.26EE40 pKa = 3.74HH41 pKa = 7.2APTTGVPHH49 pKa = 6.69LQGFCNLSKK58 pKa = 10.66PMRR61 pKa = 11.84FSAIKK66 pKa = 9.89KK67 pKa = 8.67CLSNSIHH74 pKa = 6.31IEE76 pKa = 3.76KK77 pKa = 10.55AIGSDD82 pKa = 3.42EE83 pKa = 4.04QNQKK87 pKa = 10.03YY88 pKa = 9.71CSKK91 pKa = 10.96AGDD94 pKa = 3.8FFEE97 pKa = 5.43EE98 pKa = 4.57GLPYY102 pKa = 9.93TQGKK106 pKa = 7.77RR107 pKa = 11.84TDD109 pKa = 3.5LEE111 pKa = 4.07AVVRR115 pKa = 11.84DD116 pKa = 3.94MEE118 pKa = 4.35QGTVRR123 pKa = 11.84LEE125 pKa = 4.59DD126 pKa = 3.15IAAKK130 pKa = 10.13HH131 pKa = 5.52PCAFIKK137 pKa = 8.34YY138 pKa = 9.62HH139 pKa = 6.87RR140 pKa = 11.84GIDD143 pKa = 3.5QLLKK147 pKa = 10.34LIHH150 pKa = 6.8PIKK153 pKa = 10.54PRR155 pKa = 11.84DD156 pKa = 3.56FQTYY160 pKa = 7.78VYY162 pKa = 9.12YY163 pKa = 10.77YY164 pKa = 8.96WGEE167 pKa = 4.08PGTGKK172 pKa = 10.02SRR174 pKa = 11.84RR175 pKa = 11.84AKK177 pKa = 10.62SEE179 pKa = 3.58ADD181 pKa = 3.28AVNSASIYY189 pKa = 8.73YY190 pKa = 9.87KK191 pKa = 10.51PRR193 pKa = 11.84GLWWDD198 pKa = 4.4GYY200 pKa = 9.51RR201 pKa = 11.84QQEE204 pKa = 4.04NVIIDD209 pKa = 4.38DD210 pKa = 4.37FYY212 pKa = 11.73GWIKK216 pKa = 10.75YY217 pKa = 10.28DD218 pKa = 4.1EE219 pKa = 4.44LLKK222 pKa = 10.71ICDD225 pKa = 3.53RR226 pKa = 11.84YY227 pKa = 9.88PYY229 pKa = 10.1KK230 pKa = 10.94VQIKK234 pKa = 10.43GGFEE238 pKa = 3.92EE239 pKa = 4.93FTSKK243 pKa = 10.88RR244 pKa = 11.84IWITSNVDD252 pKa = 2.54IYY254 pKa = 11.62SLYY257 pKa = 10.75KK258 pKa = 10.27FEE260 pKa = 5.17GYY262 pKa = 9.61NAEE265 pKa = 3.93AFKK268 pKa = 10.98RR269 pKa = 11.84RR270 pKa = 11.84ITCMVEE276 pKa = 3.78MNSNKK281 pKa = 10.17
Molecular weight: 32.86 kDa
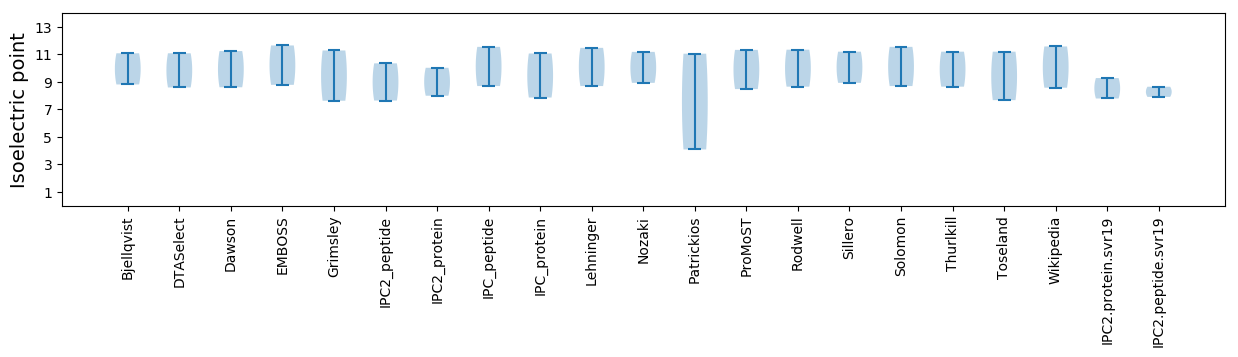
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D9IZ81|D9IZ81_9CIRC Putative Cap OS=Bat cyclovirus GF-4c OX=795381 PE=4 SV=1
MM1 pKa = 7.28AAFRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84VRR11 pKa = 11.84PRR13 pKa = 11.84KK14 pKa = 9.71NRR16 pKa = 11.84FGRR19 pKa = 11.84LRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84FGRR27 pKa = 11.84RR28 pKa = 11.84IGRR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 9.34VRR35 pKa = 11.84PNVLYY40 pKa = 10.73CKK42 pKa = 9.8LTRR45 pKa = 11.84TVTRR49 pKa = 11.84KK50 pKa = 9.13IDD52 pKa = 3.34LRR54 pKa = 11.84IATGSPNQIEE64 pKa = 4.25IQEE67 pKa = 3.97LRR69 pKa = 11.84INLNNFAEE77 pKa = 4.91HH78 pKa = 6.87INLASNFEE86 pKa = 4.37KK87 pKa = 10.53IKK89 pKa = 10.37ILKK92 pKa = 9.49QVVSVIPCQNVSNSTTSIQLHH113 pKa = 5.31YY114 pKa = 10.87AIVPYY119 pKa = 10.37KK120 pKa = 10.33KK121 pKa = 10.12EE122 pKa = 4.12SPALSTPFPAILSVDD137 pKa = 3.49KK138 pKa = 11.33AKK140 pKa = 10.36MISNTRR146 pKa = 11.84RR147 pKa = 11.84ASMALVPAAHH157 pKa = 6.66LHH159 pKa = 6.22SSGQTSDD166 pKa = 4.53DD167 pKa = 3.75MNKK170 pKa = 10.13LVYY173 pKa = 10.08KK174 pKa = 10.28PEE176 pKa = 3.86FSIPATGVSATIYY189 pKa = 9.83SGFVAFEE196 pKa = 4.21RR197 pKa = 11.84NSDD200 pKa = 3.53EE201 pKa = 4.48VFEE204 pKa = 5.15NKK206 pKa = 9.57QAWFVVKK213 pKa = 10.14QDD215 pKa = 3.28IYY217 pKa = 11.58VKK219 pKa = 10.75YY220 pKa = 9.75SVQRR224 pKa = 11.84SFII227 pKa = 3.98
MM1 pKa = 7.28AAFRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84VRR11 pKa = 11.84PRR13 pKa = 11.84KK14 pKa = 9.71NRR16 pKa = 11.84FGRR19 pKa = 11.84LRR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84FGRR27 pKa = 11.84RR28 pKa = 11.84IGRR31 pKa = 11.84RR32 pKa = 11.84KK33 pKa = 9.34VRR35 pKa = 11.84PNVLYY40 pKa = 10.73CKK42 pKa = 9.8LTRR45 pKa = 11.84TVTRR49 pKa = 11.84KK50 pKa = 9.13IDD52 pKa = 3.34LRR54 pKa = 11.84IATGSPNQIEE64 pKa = 4.25IQEE67 pKa = 3.97LRR69 pKa = 11.84INLNNFAEE77 pKa = 4.91HH78 pKa = 6.87INLASNFEE86 pKa = 4.37KK87 pKa = 10.53IKK89 pKa = 10.37ILKK92 pKa = 9.49QVVSVIPCQNVSNSTTSIQLHH113 pKa = 5.31YY114 pKa = 10.87AIVPYY119 pKa = 10.37KK120 pKa = 10.33KK121 pKa = 10.12EE122 pKa = 4.12SPALSTPFPAILSVDD137 pKa = 3.49KK138 pKa = 11.33AKK140 pKa = 10.36MISNTRR146 pKa = 11.84RR147 pKa = 11.84ASMALVPAAHH157 pKa = 6.66LHH159 pKa = 6.22SSGQTSDD166 pKa = 4.53DD167 pKa = 3.75MNKK170 pKa = 10.13LVYY173 pKa = 10.08KK174 pKa = 10.28PEE176 pKa = 3.86FSIPATGVSATIYY189 pKa = 9.83SGFVAFEE196 pKa = 4.21RR197 pKa = 11.84NSDD200 pKa = 3.53EE201 pKa = 4.48VFEE204 pKa = 5.15NKK206 pKa = 9.57QAWFVVKK213 pKa = 10.14QDD215 pKa = 3.28IYY217 pKa = 11.58VKK219 pKa = 10.75YY220 pKa = 9.75SVQRR224 pKa = 11.84SFII227 pKa = 3.98
Molecular weight: 26.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
508 |
227 |
281 |
254.0 |
29.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.299 ± 0.726 | 1.969 ± 0.663 |
4.724 ± 1.269 | 5.315 ± 0.823 |
5.118 ± 0.103 | 5.315 ± 1.361 |
1.969 ± 0.126 | 8.071 ± 0.086 |
8.465 ± 0.864 | 5.315 ± 0.52 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.772 ± 0.006 | 5.315 ± 0.52 |
4.134 ± 0.434 | 3.74 ± 0.137 |
8.268 ± 1.943 | 6.693 ± 1.291 |
4.921 ± 0.046 | 5.906 ± 1.503 |
1.378 ± 0.572 | 5.315 ± 1.361 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
