
Hubei tombus-like virus 18
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.98
Get precalculated fractions of proteins
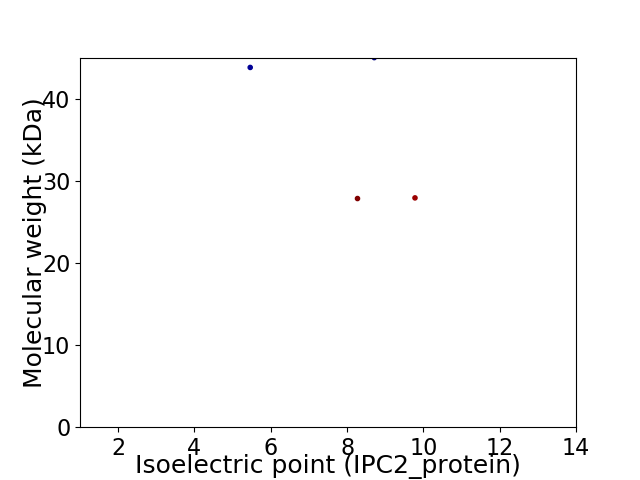
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KG77|A0A1L3KG77_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 18 OX=1923264 PE=4 SV=1
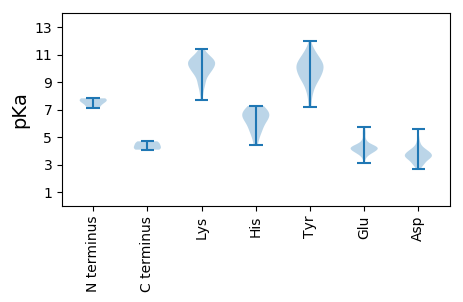
MM1 pKa = 7.71ANAQPAQGIVGLEE14 pKa = 3.84NIIAQLQQLTQDD26 pKa = 4.06PNFDD30 pKa = 3.65PQLAEE35 pKa = 4.35DD36 pKa = 5.59LILATRR42 pKa = 11.84PVLEE46 pKa = 3.75AHH48 pKa = 6.66RR49 pKa = 11.84LRR51 pKa = 11.84RR52 pKa = 11.84EE53 pKa = 3.95RR54 pKa = 11.84VVLARR59 pKa = 11.84DD60 pKa = 3.55LLIALQDD67 pKa = 4.09GEE69 pKa = 5.29DD70 pKa = 3.72PAGDD74 pKa = 3.62GLGIPLVADD83 pKa = 3.49ALEE86 pKa = 4.27NVQRR90 pKa = 11.84EE91 pKa = 4.4IIRR94 pKa = 11.84EE95 pKa = 3.99RR96 pKa = 11.84GHH98 pKa = 6.6PGVPGGGMHH107 pKa = 7.26RR108 pKa = 11.84DD109 pKa = 3.0AGLFVGEE116 pKa = 4.2GNGARR121 pKa = 11.84EE122 pKa = 4.06RR123 pKa = 11.84APNDD127 pKa = 2.98AAAEE131 pKa = 3.99LRR133 pKa = 11.84DD134 pKa = 3.48LRR136 pKa = 11.84VVRR139 pKa = 11.84EE140 pKa = 3.96RR141 pKa = 11.84DD142 pKa = 3.02EE143 pKa = 4.34HH144 pKa = 6.11RR145 pKa = 11.84RR146 pKa = 11.84QLAEE150 pKa = 4.09LPDD153 pKa = 3.69NEE155 pKa = 5.24EE156 pKa = 3.73IFGDD160 pKa = 3.96VWWMFGDD167 pKa = 4.05EE168 pKa = 4.47EE169 pKa = 4.29PAAEE173 pKa = 4.26PPAGRR178 pKa = 11.84EE179 pKa = 3.76IAVGPDD185 pKa = 2.98IPGTAVPAKK194 pKa = 10.59VRR196 pKa = 11.84DD197 pKa = 3.8KK198 pKa = 11.18YY199 pKa = 10.98RR200 pKa = 11.84SNKK203 pKa = 8.43TDD205 pKa = 2.76IRR207 pKa = 11.84DD208 pKa = 3.98PLPEE212 pKa = 3.93ATRR215 pKa = 11.84DD216 pKa = 3.69LPEE219 pKa = 4.01FRR221 pKa = 11.84ADD223 pKa = 3.51LVSVQAGTVPLFTHH237 pKa = 6.79RR238 pKa = 11.84VIDD241 pKa = 4.09HH242 pKa = 7.07RR243 pKa = 11.84PLCDD247 pKa = 3.17QDD249 pKa = 4.99VYY251 pKa = 12.0AHH253 pKa = 6.38LQAVTMFMPRR263 pKa = 11.84TPGLAVYY270 pKa = 9.14MRR272 pKa = 11.84EE273 pKa = 3.58RR274 pKa = 11.84ALRR277 pKa = 11.84FVEE280 pKa = 5.73GYY282 pKa = 8.04DD283 pKa = 3.2TGRR286 pKa = 11.84LSRR289 pKa = 11.84MEE291 pKa = 3.81INEE294 pKa = 4.27IIGAAIANAYY304 pKa = 8.49VPSVAEE310 pKa = 3.8MQLIRR315 pKa = 11.84TAEE318 pKa = 4.28YY319 pKa = 10.1IPTQYY324 pKa = 11.23RR325 pKa = 11.84LDD327 pKa = 3.62RR328 pKa = 11.84FNEE331 pKa = 4.23FFRR334 pKa = 11.84TGDD337 pKa = 3.55SSSSAATGPKK347 pKa = 10.1NSLNKK352 pKa = 9.61VNVVRR357 pKa = 11.84KK358 pKa = 9.28CLSKK362 pKa = 10.54RR363 pKa = 11.84WIVGIGLLAMASLVILRR380 pKa = 11.84KK381 pKa = 8.9PQLPLHH387 pKa = 6.55IPTLSRR393 pKa = 11.84IMM395 pKa = 4.08
MM1 pKa = 7.71ANAQPAQGIVGLEE14 pKa = 3.84NIIAQLQQLTQDD26 pKa = 4.06PNFDD30 pKa = 3.65PQLAEE35 pKa = 4.35DD36 pKa = 5.59LILATRR42 pKa = 11.84PVLEE46 pKa = 3.75AHH48 pKa = 6.66RR49 pKa = 11.84LRR51 pKa = 11.84RR52 pKa = 11.84EE53 pKa = 3.95RR54 pKa = 11.84VVLARR59 pKa = 11.84DD60 pKa = 3.55LLIALQDD67 pKa = 4.09GEE69 pKa = 5.29DD70 pKa = 3.72PAGDD74 pKa = 3.62GLGIPLVADD83 pKa = 3.49ALEE86 pKa = 4.27NVQRR90 pKa = 11.84EE91 pKa = 4.4IIRR94 pKa = 11.84EE95 pKa = 3.99RR96 pKa = 11.84GHH98 pKa = 6.6PGVPGGGMHH107 pKa = 7.26RR108 pKa = 11.84DD109 pKa = 3.0AGLFVGEE116 pKa = 4.2GNGARR121 pKa = 11.84EE122 pKa = 4.06RR123 pKa = 11.84APNDD127 pKa = 2.98AAAEE131 pKa = 3.99LRR133 pKa = 11.84DD134 pKa = 3.48LRR136 pKa = 11.84VVRR139 pKa = 11.84EE140 pKa = 3.96RR141 pKa = 11.84DD142 pKa = 3.02EE143 pKa = 4.34HH144 pKa = 6.11RR145 pKa = 11.84RR146 pKa = 11.84QLAEE150 pKa = 4.09LPDD153 pKa = 3.69NEE155 pKa = 5.24EE156 pKa = 3.73IFGDD160 pKa = 3.96VWWMFGDD167 pKa = 4.05EE168 pKa = 4.47EE169 pKa = 4.29PAAEE173 pKa = 4.26PPAGRR178 pKa = 11.84EE179 pKa = 3.76IAVGPDD185 pKa = 2.98IPGTAVPAKK194 pKa = 10.59VRR196 pKa = 11.84DD197 pKa = 3.8KK198 pKa = 11.18YY199 pKa = 10.98RR200 pKa = 11.84SNKK203 pKa = 8.43TDD205 pKa = 2.76IRR207 pKa = 11.84DD208 pKa = 3.98PLPEE212 pKa = 3.93ATRR215 pKa = 11.84DD216 pKa = 3.69LPEE219 pKa = 4.01FRR221 pKa = 11.84ADD223 pKa = 3.51LVSVQAGTVPLFTHH237 pKa = 6.79RR238 pKa = 11.84VIDD241 pKa = 4.09HH242 pKa = 7.07RR243 pKa = 11.84PLCDD247 pKa = 3.17QDD249 pKa = 4.99VYY251 pKa = 12.0AHH253 pKa = 6.38LQAVTMFMPRR263 pKa = 11.84TPGLAVYY270 pKa = 9.14MRR272 pKa = 11.84EE273 pKa = 3.58RR274 pKa = 11.84ALRR277 pKa = 11.84FVEE280 pKa = 5.73GYY282 pKa = 8.04DD283 pKa = 3.2TGRR286 pKa = 11.84LSRR289 pKa = 11.84MEE291 pKa = 3.81INEE294 pKa = 4.27IIGAAIANAYY304 pKa = 8.49VPSVAEE310 pKa = 3.8MQLIRR315 pKa = 11.84TAEE318 pKa = 4.28YY319 pKa = 10.1IPTQYY324 pKa = 11.23RR325 pKa = 11.84LDD327 pKa = 3.62RR328 pKa = 11.84FNEE331 pKa = 4.23FFRR334 pKa = 11.84TGDD337 pKa = 3.55SSSSAATGPKK347 pKa = 10.1NSLNKK352 pKa = 9.61VNVVRR357 pKa = 11.84KK358 pKa = 9.28CLSKK362 pKa = 10.54RR363 pKa = 11.84WIVGIGLLAMASLVILRR380 pKa = 11.84KK381 pKa = 8.9PQLPLHH387 pKa = 6.55IPTLSRR393 pKa = 11.84IMM395 pKa = 4.08
Molecular weight: 43.86 kDa
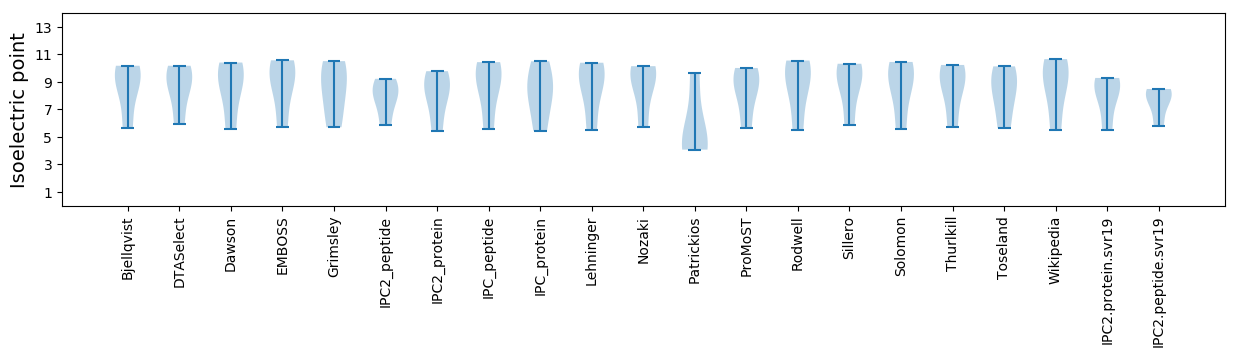
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KG35|A0A1L3KG35_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 18 OX=1923264 PE=4 SV=1
MM1 pKa = 7.13QVNRR5 pKa = 11.84YY6 pKa = 8.8LGRR9 pKa = 11.84GRR11 pKa = 11.84GWRR14 pKa = 11.84DD15 pKa = 2.7WVVPGLQVGGYY26 pKa = 8.5LANSAIRR33 pKa = 11.84SYY35 pKa = 11.02LGRR38 pKa = 11.84PQTIVYY44 pKa = 7.36QQPDD48 pKa = 3.73PLASQMANMSIQPAQQSRR66 pKa = 11.84ARR68 pKa = 11.84GGRR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84GRR75 pKa = 11.84GGRR78 pKa = 11.84GRR80 pKa = 11.84GGRR83 pKa = 11.84GRR85 pKa = 11.84GRR87 pKa = 11.84GALPLIAPPGSGAIVAEE104 pKa = 4.17DD105 pKa = 3.74TEE107 pKa = 4.76IIVVKK112 pKa = 9.31TADD115 pKa = 3.48KK116 pKa = 9.44PAVVALVFNPAVTEE130 pKa = 4.22TPRR133 pKa = 11.84LLRR136 pKa = 11.84YY137 pKa = 9.81EE138 pKa = 4.31EE139 pKa = 3.84MFEE142 pKa = 4.24RR143 pKa = 11.84YY144 pKa = 8.63RR145 pKa = 11.84VVYY148 pKa = 9.27FRR150 pKa = 11.84INYY153 pKa = 9.43KK154 pKa = 9.85GACSANQRR162 pKa = 11.84GNIQFAITPEE172 pKa = 4.03AKK174 pKa = 9.93SANVTVDD181 pKa = 4.71KK182 pKa = 10.74IAQLKK187 pKa = 8.98PYY189 pKa = 10.09RR190 pKa = 11.84SVACWQTGSIKK201 pKa = 9.83VDD203 pKa = 3.13QRR205 pKa = 11.84VGEE208 pKa = 4.18TRR210 pKa = 11.84YY211 pKa = 10.26LEE213 pKa = 4.61CGGSQPAFTFYY224 pKa = 10.93YY225 pKa = 9.4VASGEE230 pKa = 4.15EE231 pKa = 4.37GQVVGNFHH239 pKa = 6.53VSYY242 pKa = 9.85KK243 pKa = 10.4IQFAYY248 pKa = 9.83PKK250 pKa = 10.58VFTT253 pKa = 4.69
MM1 pKa = 7.13QVNRR5 pKa = 11.84YY6 pKa = 8.8LGRR9 pKa = 11.84GRR11 pKa = 11.84GWRR14 pKa = 11.84DD15 pKa = 2.7WVVPGLQVGGYY26 pKa = 8.5LANSAIRR33 pKa = 11.84SYY35 pKa = 11.02LGRR38 pKa = 11.84PQTIVYY44 pKa = 7.36QQPDD48 pKa = 3.73PLASQMANMSIQPAQQSRR66 pKa = 11.84ARR68 pKa = 11.84GGRR71 pKa = 11.84RR72 pKa = 11.84RR73 pKa = 11.84GRR75 pKa = 11.84GGRR78 pKa = 11.84GRR80 pKa = 11.84GGRR83 pKa = 11.84GRR85 pKa = 11.84GRR87 pKa = 11.84GALPLIAPPGSGAIVAEE104 pKa = 4.17DD105 pKa = 3.74TEE107 pKa = 4.76IIVVKK112 pKa = 9.31TADD115 pKa = 3.48KK116 pKa = 9.44PAVVALVFNPAVTEE130 pKa = 4.22TPRR133 pKa = 11.84LLRR136 pKa = 11.84YY137 pKa = 9.81EE138 pKa = 4.31EE139 pKa = 3.84MFEE142 pKa = 4.24RR143 pKa = 11.84YY144 pKa = 8.63RR145 pKa = 11.84VVYY148 pKa = 9.27FRR150 pKa = 11.84INYY153 pKa = 9.43KK154 pKa = 9.85GACSANQRR162 pKa = 11.84GNIQFAITPEE172 pKa = 4.03AKK174 pKa = 9.93SANVTVDD181 pKa = 4.71KK182 pKa = 10.74IAQLKK187 pKa = 8.98PYY189 pKa = 10.09RR190 pKa = 11.84SVACWQTGSIKK201 pKa = 9.83VDD203 pKa = 3.13QRR205 pKa = 11.84VGEE208 pKa = 4.18TRR210 pKa = 11.84YY211 pKa = 10.26LEE213 pKa = 4.61CGGSQPAFTFYY224 pKa = 10.93YY225 pKa = 9.4VASGEE230 pKa = 4.15EE231 pKa = 4.37GQVVGNFHH239 pKa = 6.53VSYY242 pKa = 9.85KK243 pKa = 10.4IQFAYY248 pKa = 9.83PKK250 pKa = 10.58VFTT253 pKa = 4.69
Molecular weight: 27.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1279 |
241 |
395 |
319.8 |
36.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.74 ± 1.252 | 2.033 ± 0.652 |
4.222 ± 1.103 | 6.099 ± 0.65 |
3.362 ± 0.642 | 7.506 ± 1.173 |
1.486 ± 0.46 | 5.317 ± 0.345 |
5.16 ± 1.373 | 9.851 ± 1.647 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.049 ± 0.754 | 3.909 ± 0.342 |
5.317 ± 0.809 | 4.926 ± 1.06 |
8.21 ± 0.997 | 5.16 ± 0.749 |
4.378 ± 0.493 | 6.724 ± 0.845 |
1.486 ± 0.313 | 4.066 ± 0.798 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
