
Hubei sobemo-like virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.62
Get precalculated fractions of proteins
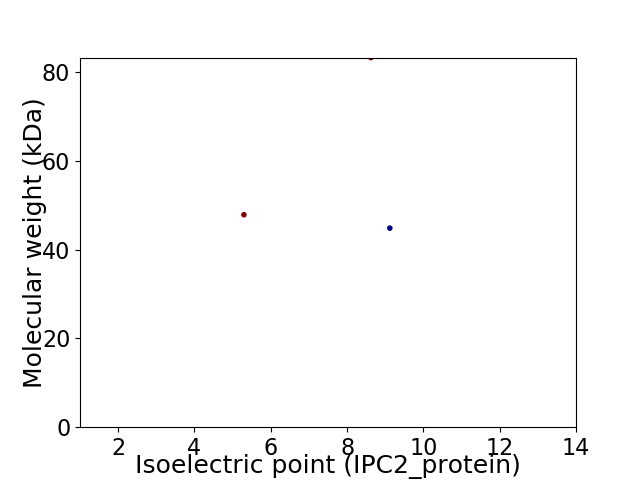
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
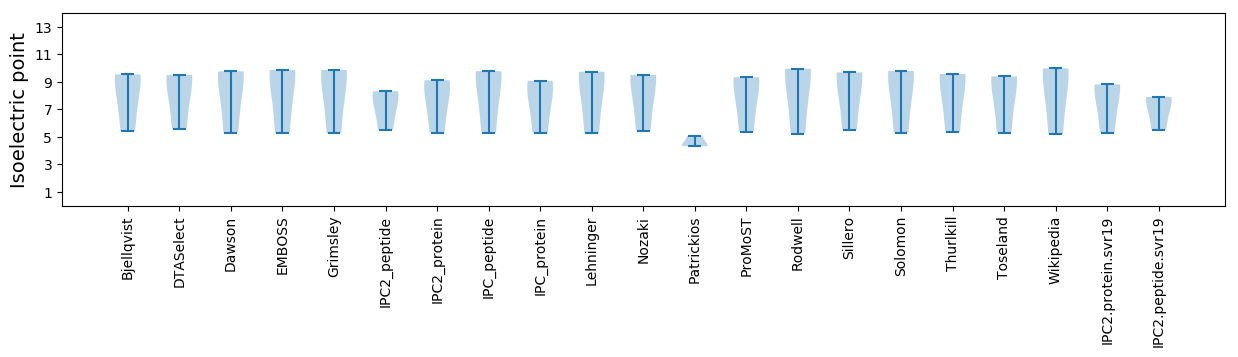
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KF00|A0A1L3KF00_9VIRU Capsid protein OS=Hubei sobemo-like virus 3 OX=1923216 PE=3 SV=1
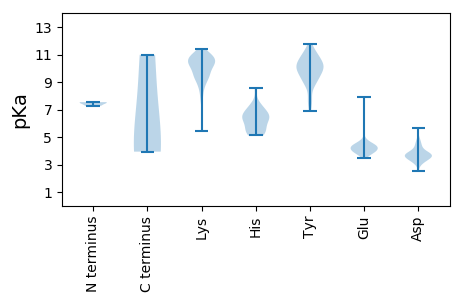
MM1 pKa = 7.54EE2 pKa = 5.68GDD4 pKa = 4.04DD5 pKa = 4.66VKK7 pKa = 11.3EE8 pKa = 4.35CSTKK12 pKa = 10.01VAEE15 pKa = 4.66MYY17 pKa = 10.03QAAGVAWDD25 pKa = 3.65FAFRR29 pKa = 11.84SYY31 pKa = 9.63EE32 pKa = 3.93TFAEE36 pKa = 4.29DD37 pKa = 4.49FEE39 pKa = 4.52NAYY42 pKa = 9.9EE43 pKa = 4.83SNVCHH48 pKa = 6.92IKK50 pKa = 8.69PTSGPGYY57 pKa = 9.14PYY59 pKa = 11.08RR60 pKa = 11.84LFGDD64 pKa = 3.73EE65 pKa = 3.84NRR67 pKa = 11.84EE68 pKa = 3.8ILEE71 pKa = 4.06SHH73 pKa = 5.82GAEE76 pKa = 3.87IKK78 pKa = 10.64RR79 pKa = 11.84LTKK82 pKa = 9.69MRR84 pKa = 11.84LEE86 pKa = 4.45TILFGGHH93 pKa = 6.34DD94 pKa = 4.14FEE96 pKa = 7.92SIAKK100 pKa = 10.34DD101 pKa = 3.25PMQWLEE107 pKa = 3.98KK108 pKa = 10.51GLRR111 pKa = 11.84DD112 pKa = 4.31PDD114 pKa = 3.42RR115 pKa = 11.84LFPKK119 pKa = 10.33NQANPLRR126 pKa = 11.84KK127 pKa = 9.14PLPRR131 pKa = 11.84VIAGSSLVDD140 pKa = 3.35QLVTRR145 pKa = 11.84ILFSGFTDD153 pKa = 3.91AEE155 pKa = 4.45GEE157 pKa = 4.31AYY159 pKa = 9.66PFLPTKK165 pKa = 10.41KK166 pKa = 10.51GIGFSDD172 pKa = 3.3EE173 pKa = 4.05HH174 pKa = 8.58AEE176 pKa = 3.89LLGEE180 pKa = 4.2QFEE183 pKa = 4.89GLNKK187 pKa = 10.5ALGRR191 pKa = 11.84PPAVSDD197 pKa = 3.47VAGWEE202 pKa = 4.4KK203 pKa = 11.02NFSEE207 pKa = 4.65PVAEE211 pKa = 4.53CTRR214 pKa = 11.84IPMKK218 pKa = 10.07EE219 pKa = 4.14TMKK222 pKa = 10.75SGSKK226 pKa = 10.21EE227 pKa = 3.83LFDD230 pKa = 5.66VGFNWWKK237 pKa = 10.55FSLLSNLAVTDD248 pKa = 3.2SGKK251 pKa = 10.39LVRR254 pKa = 11.84FKK256 pKa = 10.81DD257 pKa = 3.64LKK259 pKa = 9.58VQRR262 pKa = 11.84SGNFLTTTSNGIGRR276 pKa = 11.84KK277 pKa = 8.89CVALSVGSVANTAGDD292 pKa = 4.59DD293 pKa = 3.7CHH295 pKa = 6.58EE296 pKa = 4.34WNDD299 pKa = 3.63LSVEE303 pKa = 4.11EE304 pKa = 5.47LIQAYY309 pKa = 10.04ARR311 pKa = 11.84IGVPVRR317 pKa = 11.84DD318 pKa = 3.94VVQMDD323 pKa = 3.57SKK325 pKa = 10.0TLVFCSHH332 pKa = 6.24SFQRR336 pKa = 11.84DD337 pKa = 2.55GDD339 pKa = 4.18GRR341 pKa = 11.84WKK343 pKa = 10.43CWLSEE348 pKa = 4.19WEE350 pKa = 4.06RR351 pKa = 11.84MLYY354 pKa = 9.34EE355 pKa = 4.32ASRR358 pKa = 11.84SKK360 pKa = 11.06LLDD363 pKa = 3.24VGTDD367 pKa = 3.6LNWIKK372 pKa = 10.4EE373 pKa = 4.44VEE375 pKa = 4.13NHH377 pKa = 7.25PDD379 pKa = 3.07PEE381 pKa = 4.14MRR383 pKa = 11.84RR384 pKa = 11.84KK385 pKa = 9.56FHH387 pKa = 6.69AFVSGRR393 pKa = 11.84RR394 pKa = 11.84LLLGAVAEE402 pKa = 4.12HH403 pKa = 7.34DD404 pKa = 3.95EE405 pKa = 4.33VSEE408 pKa = 4.46SGSSHH413 pKa = 6.53PAEE416 pKa = 4.47KK417 pKa = 10.81GCAKK421 pKa = 10.35RR422 pKa = 11.84NEE424 pKa = 4.36SEE426 pKa = 3.94
MM1 pKa = 7.54EE2 pKa = 5.68GDD4 pKa = 4.04DD5 pKa = 4.66VKK7 pKa = 11.3EE8 pKa = 4.35CSTKK12 pKa = 10.01VAEE15 pKa = 4.66MYY17 pKa = 10.03QAAGVAWDD25 pKa = 3.65FAFRR29 pKa = 11.84SYY31 pKa = 9.63EE32 pKa = 3.93TFAEE36 pKa = 4.29DD37 pKa = 4.49FEE39 pKa = 4.52NAYY42 pKa = 9.9EE43 pKa = 4.83SNVCHH48 pKa = 6.92IKK50 pKa = 8.69PTSGPGYY57 pKa = 9.14PYY59 pKa = 11.08RR60 pKa = 11.84LFGDD64 pKa = 3.73EE65 pKa = 3.84NRR67 pKa = 11.84EE68 pKa = 3.8ILEE71 pKa = 4.06SHH73 pKa = 5.82GAEE76 pKa = 3.87IKK78 pKa = 10.64RR79 pKa = 11.84LTKK82 pKa = 9.69MRR84 pKa = 11.84LEE86 pKa = 4.45TILFGGHH93 pKa = 6.34DD94 pKa = 4.14FEE96 pKa = 7.92SIAKK100 pKa = 10.34DD101 pKa = 3.25PMQWLEE107 pKa = 3.98KK108 pKa = 10.51GLRR111 pKa = 11.84DD112 pKa = 4.31PDD114 pKa = 3.42RR115 pKa = 11.84LFPKK119 pKa = 10.33NQANPLRR126 pKa = 11.84KK127 pKa = 9.14PLPRR131 pKa = 11.84VIAGSSLVDD140 pKa = 3.35QLVTRR145 pKa = 11.84ILFSGFTDD153 pKa = 3.91AEE155 pKa = 4.45GEE157 pKa = 4.31AYY159 pKa = 9.66PFLPTKK165 pKa = 10.41KK166 pKa = 10.51GIGFSDD172 pKa = 3.3EE173 pKa = 4.05HH174 pKa = 8.58AEE176 pKa = 3.89LLGEE180 pKa = 4.2QFEE183 pKa = 4.89GLNKK187 pKa = 10.5ALGRR191 pKa = 11.84PPAVSDD197 pKa = 3.47VAGWEE202 pKa = 4.4KK203 pKa = 11.02NFSEE207 pKa = 4.65PVAEE211 pKa = 4.53CTRR214 pKa = 11.84IPMKK218 pKa = 10.07EE219 pKa = 4.14TMKK222 pKa = 10.75SGSKK226 pKa = 10.21EE227 pKa = 3.83LFDD230 pKa = 5.66VGFNWWKK237 pKa = 10.55FSLLSNLAVTDD248 pKa = 3.2SGKK251 pKa = 10.39LVRR254 pKa = 11.84FKK256 pKa = 10.81DD257 pKa = 3.64LKK259 pKa = 9.58VQRR262 pKa = 11.84SGNFLTTTSNGIGRR276 pKa = 11.84KK277 pKa = 8.89CVALSVGSVANTAGDD292 pKa = 4.59DD293 pKa = 3.7CHH295 pKa = 6.58EE296 pKa = 4.34WNDD299 pKa = 3.63LSVEE303 pKa = 4.11EE304 pKa = 5.47LIQAYY309 pKa = 10.04ARR311 pKa = 11.84IGVPVRR317 pKa = 11.84DD318 pKa = 3.94VVQMDD323 pKa = 3.57SKK325 pKa = 10.0TLVFCSHH332 pKa = 6.24SFQRR336 pKa = 11.84DD337 pKa = 2.55GDD339 pKa = 4.18GRR341 pKa = 11.84WKK343 pKa = 10.43CWLSEE348 pKa = 4.19WEE350 pKa = 4.06RR351 pKa = 11.84MLYY354 pKa = 9.34EE355 pKa = 4.32ASRR358 pKa = 11.84SKK360 pKa = 11.06LLDD363 pKa = 3.24VGTDD367 pKa = 3.6LNWIKK372 pKa = 10.4EE373 pKa = 4.44VEE375 pKa = 4.13NHH377 pKa = 7.25PDD379 pKa = 3.07PEE381 pKa = 4.14MRR383 pKa = 11.84RR384 pKa = 11.84KK385 pKa = 9.56FHH387 pKa = 6.69AFVSGRR393 pKa = 11.84RR394 pKa = 11.84LLLGAVAEE402 pKa = 4.12HH403 pKa = 7.34DD404 pKa = 3.95EE405 pKa = 4.33VSEE408 pKa = 4.46SGSSHH413 pKa = 6.53PAEE416 pKa = 4.47KK417 pKa = 10.81GCAKK421 pKa = 10.35RR422 pKa = 11.84NEE424 pKa = 4.36SEE426 pKa = 3.94
Molecular weight: 47.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KF02|A0A1L3KF02_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 3 OX=1923216 PE=4 SV=1
MM1 pKa = 7.27TKK3 pKa = 10.12SANRR7 pKa = 11.84GAATPQKK14 pKa = 10.29KK15 pKa = 9.38AAQRR19 pKa = 11.84GMKK22 pKa = 9.8VSNKK26 pKa = 9.24PNPIRR31 pKa = 11.84KK32 pKa = 7.86KK33 pKa = 8.72QKK35 pKa = 7.69RR36 pKa = 11.84TRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84NRR42 pKa = 11.84PAQVGLAYY50 pKa = 9.51ATGSSAKK57 pKa = 7.63MTSIVVEE64 pKa = 3.96KK65 pKa = 9.56TEE67 pKa = 3.94RR68 pKa = 11.84LGTIVSPATASGVVLTQYY86 pKa = 11.12VNAGNLALSTSSYY99 pKa = 10.67LGRR102 pKa = 11.84QAALFDD108 pKa = 4.2KK109 pKa = 10.82YY110 pKa = 10.83QFSKK114 pKa = 11.37FEE116 pKa = 3.88IEE118 pKa = 4.11YY119 pKa = 10.39VPIVSASTSGNVIIGMDD136 pKa = 4.6LSPDD140 pKa = 3.44DD141 pKa = 4.87AAPGDD146 pKa = 3.89ASGMTNLSLGYY157 pKa = 10.59AEE159 pKa = 4.49GNAWRR164 pKa = 11.84NFKK167 pKa = 10.44YY168 pKa = 10.21AAQCFACFPAGPKK181 pKa = 9.82FVRR184 pKa = 11.84STTEE188 pKa = 3.44QLGTNASLFDD198 pKa = 3.78MGSLYY203 pKa = 10.31IFTEE207 pKa = 4.68GAPVNTTLGYY217 pKa = 10.04IDD219 pKa = 3.4VHH221 pKa = 6.51YY222 pKa = 10.1KK223 pKa = 9.78VHH225 pKa = 5.71LHH227 pKa = 5.3GVNRR231 pKa = 11.84NANDD235 pKa = 3.4SPAGLLRR242 pKa = 11.84PAALATLSPTSTSAIVPGGTYY263 pKa = 9.01ATTTFASFRR272 pKa = 11.84GFSPSGIGQSDD283 pKa = 3.93MVFSAVTPAEE293 pKa = 4.24VYY295 pKa = 9.19GTVGNYY301 pKa = 6.92ITYY304 pKa = 7.63GTNSITLNAGTYY316 pKa = 7.75SVKK319 pKa = 9.84LTSCVCSDD327 pKa = 3.83TYY329 pKa = 11.79SNGLVRR335 pKa = 11.84LKK337 pKa = 10.67RR338 pKa = 11.84DD339 pKa = 3.5GNVLLSNAHH348 pKa = 5.34GWGSLSLPTTAYY360 pKa = 9.85VQTGSVDD367 pKa = 3.57SIEE370 pKa = 4.58RR371 pKa = 11.84SFTVTTTSTLTLDD384 pKa = 3.32TFVFYY389 pKa = 10.92SATPYY394 pKa = 8.76TQNKK398 pKa = 5.42TWNVVYY404 pKa = 11.02VNGDD408 pKa = 3.73SVPSTFITLTRR419 pKa = 11.84LAAA422 pKa = 4.34
MM1 pKa = 7.27TKK3 pKa = 10.12SANRR7 pKa = 11.84GAATPQKK14 pKa = 10.29KK15 pKa = 9.38AAQRR19 pKa = 11.84GMKK22 pKa = 9.8VSNKK26 pKa = 9.24PNPIRR31 pKa = 11.84KK32 pKa = 7.86KK33 pKa = 8.72QKK35 pKa = 7.69RR36 pKa = 11.84TRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84NRR42 pKa = 11.84PAQVGLAYY50 pKa = 9.51ATGSSAKK57 pKa = 7.63MTSIVVEE64 pKa = 3.96KK65 pKa = 9.56TEE67 pKa = 3.94RR68 pKa = 11.84LGTIVSPATASGVVLTQYY86 pKa = 11.12VNAGNLALSTSSYY99 pKa = 10.67LGRR102 pKa = 11.84QAALFDD108 pKa = 4.2KK109 pKa = 10.82YY110 pKa = 10.83QFSKK114 pKa = 11.37FEE116 pKa = 3.88IEE118 pKa = 4.11YY119 pKa = 10.39VPIVSASTSGNVIIGMDD136 pKa = 4.6LSPDD140 pKa = 3.44DD141 pKa = 4.87AAPGDD146 pKa = 3.89ASGMTNLSLGYY157 pKa = 10.59AEE159 pKa = 4.49GNAWRR164 pKa = 11.84NFKK167 pKa = 10.44YY168 pKa = 10.21AAQCFACFPAGPKK181 pKa = 9.82FVRR184 pKa = 11.84STTEE188 pKa = 3.44QLGTNASLFDD198 pKa = 3.78MGSLYY203 pKa = 10.31IFTEE207 pKa = 4.68GAPVNTTLGYY217 pKa = 10.04IDD219 pKa = 3.4VHH221 pKa = 6.51YY222 pKa = 10.1KK223 pKa = 9.78VHH225 pKa = 5.71LHH227 pKa = 5.3GVNRR231 pKa = 11.84NANDD235 pKa = 3.4SPAGLLRR242 pKa = 11.84PAALATLSPTSTSAIVPGGTYY263 pKa = 9.01ATTTFASFRR272 pKa = 11.84GFSPSGIGQSDD283 pKa = 3.93MVFSAVTPAEE293 pKa = 4.24VYY295 pKa = 9.19GTVGNYY301 pKa = 6.92ITYY304 pKa = 7.63GTNSITLNAGTYY316 pKa = 7.75SVKK319 pKa = 9.84LTSCVCSDD327 pKa = 3.83TYY329 pKa = 11.79SNGLVRR335 pKa = 11.84LKK337 pKa = 10.67RR338 pKa = 11.84DD339 pKa = 3.5GNVLLSNAHH348 pKa = 5.34GWGSLSLPTTAYY360 pKa = 9.85VQTGSVDD367 pKa = 3.57SIEE370 pKa = 4.58RR371 pKa = 11.84SFTVTTTSTLTLDD384 pKa = 3.32TFVFYY389 pKa = 10.92SATPYY394 pKa = 8.76TQNKK398 pKa = 5.42TWNVVYY404 pKa = 11.02VNGDD408 pKa = 3.73SVPSTFITLTRR419 pKa = 11.84LAAA422 pKa = 4.34
Molecular weight: 44.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1618 |
422 |
770 |
539.3 |
58.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.703 ± 0.959 | 1.422 ± 0.217 |
4.017 ± 0.83 | 5.871 ± 1.695 |
4.079 ± 0.492 | 7.54 ± 0.493 |
1.731 ± 0.335 | 3.523 ± 0.174 |
6.675 ± 1.02 | 7.417 ± 0.47 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.607 ± 0.199 | 3.708 ± 0.833 |
4.944 ± 0.225 | 3.152 ± 0.451 |
4.697 ± 0.51 | 9.642 ± 0.607 |
8.529 ± 1.782 | 7.046 ± 0.315 |
1.731 ± 0.415 | 2.967 ± 0.723 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
