
Lycianthes yellow mosaic virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Begomovirus
Average proteome isoelectric point is 8.13
Get precalculated fractions of proteins
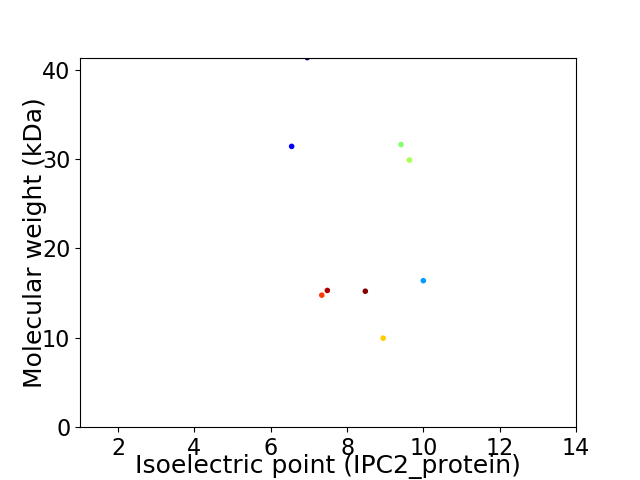
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

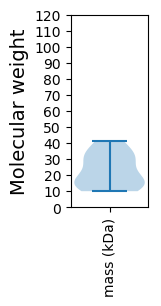
Protein with the lowest isoelectric point:
>tr|A0A140D6R5|A0A140D6R5_9GEMI Movement protein BC1 OS=Lycianthes yellow mosaic virus OX=1779714 GN=BC PE=3 SV=1
MM1 pKa = 7.33EE2 pKa = 5.69HH3 pKa = 6.59NNNAVAYY10 pKa = 5.35TTSDD14 pKa = 2.84RR15 pKa = 11.84TEE17 pKa = 3.9FQLSNEE23 pKa = 4.1KK24 pKa = 9.97TEE26 pKa = 4.14VTLLFPSTIDD36 pKa = 3.27QKK38 pKa = 11.06FSKK41 pKa = 10.79LLGKK45 pKa = 9.26CLRR48 pKa = 11.84IDD50 pKa = 3.67HH51 pKa = 6.78VILEE55 pKa = 4.31YY56 pKa = 10.8RR57 pKa = 11.84NQVPVNATGHH67 pKa = 5.14VVIEE71 pKa = 4.01MHH73 pKa = 6.5DD74 pKa = 3.69TRR76 pKa = 11.84LHH78 pKa = 6.18EE79 pKa = 4.92GDD81 pKa = 3.64SKK83 pKa = 10.99QAEE86 pKa = 4.06FTIPIGCNCNIHH98 pKa = 6.3YY99 pKa = 10.23YY100 pKa = 9.48SSSYY104 pKa = 10.47FSPKK108 pKa = 10.33DD109 pKa = 3.49PNPWRR114 pKa = 11.84VMYY117 pKa = 10.43RR118 pKa = 11.84VDD120 pKa = 3.44DD121 pKa = 4.19TNVVNGVHH129 pKa = 6.6FCRR132 pKa = 11.84MQGKK136 pKa = 9.91LKK138 pKa = 10.2MSSAKK143 pKa = 9.73QSSEE147 pKa = 3.7IQFKK151 pKa = 10.45SPKK154 pKa = 9.79IEE156 pKa = 4.1ILSKK160 pKa = 10.48AYY162 pKa = 11.2NMTNIDD168 pKa = 4.06FWTVPHH174 pKa = 6.11SQVSRR179 pKa = 11.84KK180 pKa = 8.79PVQALSNIRR189 pKa = 11.84SQSCRR194 pKa = 11.84YY195 pKa = 5.2TTPAILPGQTWASASSVGNEE215 pKa = 4.06DD216 pKa = 5.09QEE218 pKa = 4.34DD219 pKa = 3.71LPYY222 pKa = 11.02RR223 pKa = 11.84NLHH226 pKa = 5.9RR227 pKa = 11.84LQDD230 pKa = 3.65KK231 pKa = 8.97TLDD234 pKa = 4.11PGPSASEE241 pKa = 3.83VVGNNQTVNDD251 pKa = 4.22DD252 pKa = 3.87VINIIKK258 pKa = 8.83KK259 pKa = 7.36TVEE262 pKa = 3.62LCMEE266 pKa = 4.87GSNVTSNVKK275 pKa = 9.77PIKK278 pKa = 10.45
MM1 pKa = 7.33EE2 pKa = 5.69HH3 pKa = 6.59NNNAVAYY10 pKa = 5.35TTSDD14 pKa = 2.84RR15 pKa = 11.84TEE17 pKa = 3.9FQLSNEE23 pKa = 4.1KK24 pKa = 9.97TEE26 pKa = 4.14VTLLFPSTIDD36 pKa = 3.27QKK38 pKa = 11.06FSKK41 pKa = 10.79LLGKK45 pKa = 9.26CLRR48 pKa = 11.84IDD50 pKa = 3.67HH51 pKa = 6.78VILEE55 pKa = 4.31YY56 pKa = 10.8RR57 pKa = 11.84NQVPVNATGHH67 pKa = 5.14VVIEE71 pKa = 4.01MHH73 pKa = 6.5DD74 pKa = 3.69TRR76 pKa = 11.84LHH78 pKa = 6.18EE79 pKa = 4.92GDD81 pKa = 3.64SKK83 pKa = 10.99QAEE86 pKa = 4.06FTIPIGCNCNIHH98 pKa = 6.3YY99 pKa = 10.23YY100 pKa = 9.48SSSYY104 pKa = 10.47FSPKK108 pKa = 10.33DD109 pKa = 3.49PNPWRR114 pKa = 11.84VMYY117 pKa = 10.43RR118 pKa = 11.84VDD120 pKa = 3.44DD121 pKa = 4.19TNVVNGVHH129 pKa = 6.6FCRR132 pKa = 11.84MQGKK136 pKa = 9.91LKK138 pKa = 10.2MSSAKK143 pKa = 9.73QSSEE147 pKa = 3.7IQFKK151 pKa = 10.45SPKK154 pKa = 9.79IEE156 pKa = 4.1ILSKK160 pKa = 10.48AYY162 pKa = 11.2NMTNIDD168 pKa = 4.06FWTVPHH174 pKa = 6.11SQVSRR179 pKa = 11.84KK180 pKa = 8.79PVQALSNIRR189 pKa = 11.84SQSCRR194 pKa = 11.84YY195 pKa = 5.2TTPAILPGQTWASASSVGNEE215 pKa = 4.06DD216 pKa = 5.09QEE218 pKa = 4.34DD219 pKa = 3.71LPYY222 pKa = 11.02RR223 pKa = 11.84NLHH226 pKa = 5.9RR227 pKa = 11.84LQDD230 pKa = 3.65KK231 pKa = 8.97TLDD234 pKa = 4.11PGPSASEE241 pKa = 3.83VVGNNQTVNDD251 pKa = 4.22DD252 pKa = 3.87VINIIKK258 pKa = 8.83KK259 pKa = 7.36TVEE262 pKa = 3.62LCMEE266 pKa = 4.87GSNVTSNVKK275 pKa = 9.77PIKK278 pKa = 10.45
Molecular weight: 31.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140D6R2|A0A140D6R2_9GEMI AC4 protein OS=Lycianthes yellow mosaic virus OX=1779714 GN=AC4 PE=3 SV=1
MM1 pKa = 7.38AVDD4 pKa = 4.58FRR6 pKa = 11.84TGEE9 pKa = 4.38YY10 pKa = 8.66ITATQAQNGAYY21 pKa = 8.83IWTVPNPLYY30 pKa = 10.81FKK32 pKa = 10.36VLKK35 pKa = 9.04HH36 pKa = 5.45TSRR39 pKa = 11.84PFNSDD44 pKa = 2.24HH45 pKa = 7.75DD46 pKa = 4.12YY47 pKa = 11.89LEE49 pKa = 4.04IQIQFNHH56 pKa = 5.87NLRR59 pKa = 11.84RR60 pKa = 11.84ALGIHH65 pKa = 5.86KK66 pKa = 9.88CFLIFRR72 pKa = 11.84IWTRR76 pKa = 11.84LRR78 pKa = 11.84PQTWRR83 pKa = 11.84FLRR86 pKa = 11.84VFKK89 pKa = 11.21YY90 pKa = 10.66NVIRR94 pKa = 11.84FLNNLGVVSINNVIRR109 pKa = 11.84SVNHH113 pKa = 5.24VLWNVFDD120 pKa = 3.6NTTYY124 pKa = 11.61ARR126 pKa = 11.84MLYY129 pKa = 10.06AIKK132 pKa = 10.51FNIYY136 pKa = 10.14
MM1 pKa = 7.38AVDD4 pKa = 4.58FRR6 pKa = 11.84TGEE9 pKa = 4.38YY10 pKa = 8.66ITATQAQNGAYY21 pKa = 8.83IWTVPNPLYY30 pKa = 10.81FKK32 pKa = 10.36VLKK35 pKa = 9.04HH36 pKa = 5.45TSRR39 pKa = 11.84PFNSDD44 pKa = 2.24HH45 pKa = 7.75DD46 pKa = 4.12YY47 pKa = 11.89LEE49 pKa = 4.04IQIQFNHH56 pKa = 5.87NLRR59 pKa = 11.84RR60 pKa = 11.84ALGIHH65 pKa = 5.86KK66 pKa = 9.88CFLIFRR72 pKa = 11.84IWTRR76 pKa = 11.84LRR78 pKa = 11.84PQTWRR83 pKa = 11.84FLRR86 pKa = 11.84VFKK89 pKa = 11.21YY90 pKa = 10.66NVIRR94 pKa = 11.84FLNNLGVVSINNVIRR109 pKa = 11.84SVNHH113 pKa = 5.24VLWNVFDD120 pKa = 3.6NTTYY124 pKa = 11.61ARR126 pKa = 11.84MLYY129 pKa = 10.06AIKK132 pKa = 10.51FNIYY136 pKa = 10.14
Molecular weight: 16.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1798 |
85 |
362 |
199.8 |
22.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.505 ± 0.528 | 2.336 ± 0.337 |
4.672 ± 0.36 | 4.171 ± 0.551 |
4.616 ± 0.508 | 5.061 ± 0.636 |
3.782 ± 0.392 | 6.34 ± 0.687 |
5.562 ± 0.513 | 7.341 ± 0.71 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.225 ± 0.634 | 6.452 ± 0.6 |
5.006 ± 0.34 | 4.338 ± 0.543 |
6.952 ± 1.008 | 9.121 ± 0.79 |
5.228 ± 0.688 | 7.119 ± 0.788 |
1.168 ± 0.243 | 4.004 ± 0.617 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
