
Lake Sarah-associated circular virus-6
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.52
Get precalculated fractions of proteins
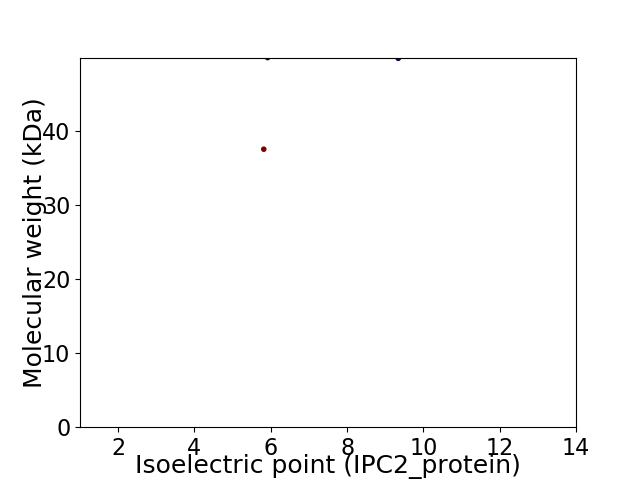
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQK0|A0A140AQK0_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-6 OX=1685783 PE=3 SV=1
MM1 pKa = 7.54EE2 pKa = 6.32PIDD5 pKa = 4.15EE6 pKa = 4.85ASQPMEE12 pKa = 5.35DD13 pKa = 4.49DD14 pKa = 4.25LGVMSPKK21 pKa = 10.41KK22 pKa = 10.27KK23 pKa = 10.3FRR25 pKa = 11.84ATAKK29 pKa = 10.37NFALTFPQCDD39 pKa = 3.65MNKK42 pKa = 9.73EE43 pKa = 3.89DD44 pKa = 4.6AFQKK48 pKa = 10.54IMDD51 pKa = 3.94EE52 pKa = 4.25HH53 pKa = 7.49KK54 pKa = 10.24PAYY57 pKa = 10.21LCVAEE62 pKa = 5.14EE63 pKa = 4.02EE64 pKa = 4.77HH65 pKa = 7.35KK66 pKa = 11.06DD67 pKa = 3.9GSPHH71 pKa = 5.42LHH73 pKa = 6.31ALISFSSKK81 pKa = 11.04KK82 pKa = 10.21NIKK85 pKa = 9.73DD86 pKa = 3.4PKK88 pKa = 10.78HH89 pKa = 6.48FDD91 pKa = 3.31LSDD94 pKa = 3.62NEE96 pKa = 4.77STFHH100 pKa = 7.44CNIQSCKK107 pKa = 10.38SVADD111 pKa = 3.42WKK113 pKa = 10.99RR114 pKa = 11.84YY115 pKa = 7.68IKK117 pKa = 10.49KK118 pKa = 10.07DD119 pKa = 3.15GKK121 pKa = 10.62YY122 pKa = 9.96KK123 pKa = 9.2EE124 pKa = 4.55TEE126 pKa = 3.81EE127 pKa = 4.98SFVLDD132 pKa = 3.63DD133 pKa = 3.88HH134 pKa = 7.68RR135 pKa = 11.84LGQRR139 pKa = 11.84QKK141 pKa = 11.11LYY143 pKa = 11.38NDD145 pKa = 4.12FCWSKK150 pKa = 9.98TYY152 pKa = 10.28IEE154 pKa = 4.17RR155 pKa = 11.84NEE157 pKa = 4.13RR158 pKa = 11.84RR159 pKa = 11.84DD160 pKa = 3.19IDD162 pKa = 3.65WPVTIRR168 pKa = 11.84DD169 pKa = 4.14SEE171 pKa = 4.39GHH173 pKa = 4.92EE174 pKa = 4.02HH175 pKa = 5.66TMEE178 pKa = 4.56RR179 pKa = 11.84PLPDD183 pKa = 3.36EE184 pKa = 4.49QGLLTKK190 pKa = 10.11KK191 pKa = 9.95RR192 pKa = 11.84NWWIVSPPNAGKK204 pKa = 8.6TYY206 pKa = 9.56WVNKK210 pKa = 6.92QFKK213 pKa = 7.99GQKK216 pKa = 9.48IFMRR220 pKa = 11.84EE221 pKa = 3.63NKK223 pKa = 9.26EE224 pKa = 4.22YY225 pKa = 10.34AWEE228 pKa = 5.02DD229 pKa = 3.67YY230 pKa = 10.95DD231 pKa = 5.08HH232 pKa = 7.34EE233 pKa = 5.61DD234 pKa = 4.58IIIVDD239 pKa = 4.12DD240 pKa = 4.09STMKK244 pKa = 10.18WEE246 pKa = 4.16EE247 pKa = 3.75LSAVCNTYY255 pKa = 10.74TNLKK259 pKa = 8.95QVPGKK264 pKa = 10.01IRR266 pKa = 11.84YY267 pKa = 8.44KK268 pKa = 10.44NLYY271 pKa = 7.96WKK273 pKa = 10.2QNHH276 pKa = 5.82ARR278 pKa = 11.84NIIILSNLKK287 pKa = 10.37PIDD290 pKa = 4.15CYY292 pKa = 11.28GQNTTILNAVLEE304 pKa = 4.35RR305 pKa = 11.84FVVLEE310 pKa = 3.91IDD312 pKa = 3.66NLINNN317 pKa = 4.66
MM1 pKa = 7.54EE2 pKa = 6.32PIDD5 pKa = 4.15EE6 pKa = 4.85ASQPMEE12 pKa = 5.35DD13 pKa = 4.49DD14 pKa = 4.25LGVMSPKK21 pKa = 10.41KK22 pKa = 10.27KK23 pKa = 10.3FRR25 pKa = 11.84ATAKK29 pKa = 10.37NFALTFPQCDD39 pKa = 3.65MNKK42 pKa = 9.73EE43 pKa = 3.89DD44 pKa = 4.6AFQKK48 pKa = 10.54IMDD51 pKa = 3.94EE52 pKa = 4.25HH53 pKa = 7.49KK54 pKa = 10.24PAYY57 pKa = 10.21LCVAEE62 pKa = 5.14EE63 pKa = 4.02EE64 pKa = 4.77HH65 pKa = 7.35KK66 pKa = 11.06DD67 pKa = 3.9GSPHH71 pKa = 5.42LHH73 pKa = 6.31ALISFSSKK81 pKa = 11.04KK82 pKa = 10.21NIKK85 pKa = 9.73DD86 pKa = 3.4PKK88 pKa = 10.78HH89 pKa = 6.48FDD91 pKa = 3.31LSDD94 pKa = 3.62NEE96 pKa = 4.77STFHH100 pKa = 7.44CNIQSCKK107 pKa = 10.38SVADD111 pKa = 3.42WKK113 pKa = 10.99RR114 pKa = 11.84YY115 pKa = 7.68IKK117 pKa = 10.49KK118 pKa = 10.07DD119 pKa = 3.15GKK121 pKa = 10.62YY122 pKa = 9.96KK123 pKa = 9.2EE124 pKa = 4.55TEE126 pKa = 3.81EE127 pKa = 4.98SFVLDD132 pKa = 3.63DD133 pKa = 3.88HH134 pKa = 7.68RR135 pKa = 11.84LGQRR139 pKa = 11.84QKK141 pKa = 11.11LYY143 pKa = 11.38NDD145 pKa = 4.12FCWSKK150 pKa = 9.98TYY152 pKa = 10.28IEE154 pKa = 4.17RR155 pKa = 11.84NEE157 pKa = 4.13RR158 pKa = 11.84RR159 pKa = 11.84DD160 pKa = 3.19IDD162 pKa = 3.65WPVTIRR168 pKa = 11.84DD169 pKa = 4.14SEE171 pKa = 4.39GHH173 pKa = 4.92EE174 pKa = 4.02HH175 pKa = 5.66TMEE178 pKa = 4.56RR179 pKa = 11.84PLPDD183 pKa = 3.36EE184 pKa = 4.49QGLLTKK190 pKa = 10.11KK191 pKa = 9.95RR192 pKa = 11.84NWWIVSPPNAGKK204 pKa = 8.6TYY206 pKa = 9.56WVNKK210 pKa = 6.92QFKK213 pKa = 7.99GQKK216 pKa = 9.48IFMRR220 pKa = 11.84EE221 pKa = 3.63NKK223 pKa = 9.26EE224 pKa = 4.22YY225 pKa = 10.34AWEE228 pKa = 5.02DD229 pKa = 3.67YY230 pKa = 10.95DD231 pKa = 5.08HH232 pKa = 7.34EE233 pKa = 5.61DD234 pKa = 4.58IIIVDD239 pKa = 4.12DD240 pKa = 4.09STMKK244 pKa = 10.18WEE246 pKa = 4.16EE247 pKa = 3.75LSAVCNTYY255 pKa = 10.74TNLKK259 pKa = 8.95QVPGKK264 pKa = 10.01IRR266 pKa = 11.84YY267 pKa = 8.44KK268 pKa = 10.44NLYY271 pKa = 7.96WKK273 pKa = 10.2QNHH276 pKa = 5.82ARR278 pKa = 11.84NIIILSNLKK287 pKa = 10.37PIDD290 pKa = 4.15CYY292 pKa = 11.28GQNTTILNAVLEE304 pKa = 4.35RR305 pKa = 11.84FVVLEE310 pKa = 3.91IDD312 pKa = 3.66NLINNN317 pKa = 4.66
Molecular weight: 37.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQK0|A0A140AQK0_9VIRU Replication associated protein OS=Lake Sarah-associated circular virus-6 OX=1685783 PE=3 SV=1
MM1 pKa = 7.47TSDD4 pKa = 3.94LQRR7 pKa = 11.84KK8 pKa = 9.22RR9 pKa = 11.84NLSSFGKK16 pKa = 10.16SFSDD20 pKa = 3.85LFMPNKK26 pKa = 9.63NKK28 pKa = 9.8RR29 pKa = 11.84RR30 pKa = 11.84NVQFGGPAKK39 pKa = 9.87RR40 pKa = 11.84AKK42 pKa = 9.88NSVVAGSFRR51 pKa = 11.84SPTVSNSAGGVIRR64 pKa = 11.84GGSQGRR70 pKa = 11.84WSSLPFQRR78 pKa = 11.84NGYY81 pKa = 8.04KK82 pKa = 10.07QYY84 pKa = 11.08INRR87 pKa = 11.84LLNPPVIITDD97 pKa = 3.2RR98 pKa = 11.84WTGLLKK104 pKa = 9.62QTVTATPDD112 pKa = 2.93LPTPVNKK119 pKa = 9.94IQNFFIDD126 pKa = 3.41SMTIGEE132 pKa = 4.44NVSAVLTNILKK143 pKa = 8.1THH145 pKa = 6.36SGHH148 pKa = 6.2GLGMQGATMTSSTFAQAILTLNTPINTVQQTFTQSFRR185 pKa = 11.84YY186 pKa = 9.28QKK188 pKa = 9.62QNKK191 pKa = 6.51VTFQNMTNNNVKK203 pKa = 8.74VTVFCFAVRR212 pKa = 11.84DD213 pKa = 3.71LRR215 pKa = 11.84TSYY218 pKa = 11.27NILTPYY224 pKa = 7.03TTLDD228 pKa = 3.93LAPGTSPNLPILPAASNAVLEE249 pKa = 4.41QAKK252 pKa = 8.04QTSGNVVQDD261 pKa = 3.82SLCVSTGFDD270 pKa = 3.59GDD272 pKa = 4.1SLNSFVAASTDD283 pKa = 3.2AQTCNWFLKK292 pKa = 10.7NPWSYY297 pKa = 11.0YY298 pKa = 7.22KK299 pKa = 10.16WRR301 pKa = 11.84MEE303 pKa = 3.81YY304 pKa = 10.43RR305 pKa = 11.84KK306 pKa = 9.94VSQVSFMLPAGQGTCTRR323 pKa = 11.84YY324 pKa = 9.68FNHH327 pKa = 6.41SFKK330 pKa = 11.23LNNKK334 pKa = 7.69WSFYY338 pKa = 8.8TEE340 pKa = 4.38WAQHH344 pKa = 5.3PAFTRR349 pKa = 11.84FYY351 pKa = 10.55LIQTEE356 pKa = 4.37TQPMSTVTTGVLPSQQGFLAYY377 pKa = 9.69PPATVGLLVEE387 pKa = 4.74SKK389 pKa = 9.56TVIQRR394 pKa = 11.84VSGVTRR400 pKa = 11.84ASHH403 pKa = 5.97LRR405 pKa = 11.84EE406 pKa = 4.08PLVDD410 pKa = 3.61PTPITGNASTGAADD424 pKa = 5.37FIAPASLVSGALNMASEE441 pKa = 4.48EE442 pKa = 4.17TDD444 pKa = 3.11QKK446 pKa = 10.32QAGVFFF452 pKa = 5.24
MM1 pKa = 7.47TSDD4 pKa = 3.94LQRR7 pKa = 11.84KK8 pKa = 9.22RR9 pKa = 11.84NLSSFGKK16 pKa = 10.16SFSDD20 pKa = 3.85LFMPNKK26 pKa = 9.63NKK28 pKa = 9.8RR29 pKa = 11.84RR30 pKa = 11.84NVQFGGPAKK39 pKa = 9.87RR40 pKa = 11.84AKK42 pKa = 9.88NSVVAGSFRR51 pKa = 11.84SPTVSNSAGGVIRR64 pKa = 11.84GGSQGRR70 pKa = 11.84WSSLPFQRR78 pKa = 11.84NGYY81 pKa = 8.04KK82 pKa = 10.07QYY84 pKa = 11.08INRR87 pKa = 11.84LLNPPVIITDD97 pKa = 3.2RR98 pKa = 11.84WTGLLKK104 pKa = 9.62QTVTATPDD112 pKa = 2.93LPTPVNKK119 pKa = 9.94IQNFFIDD126 pKa = 3.41SMTIGEE132 pKa = 4.44NVSAVLTNILKK143 pKa = 8.1THH145 pKa = 6.36SGHH148 pKa = 6.2GLGMQGATMTSSTFAQAILTLNTPINTVQQTFTQSFRR185 pKa = 11.84YY186 pKa = 9.28QKK188 pKa = 9.62QNKK191 pKa = 6.51VTFQNMTNNNVKK203 pKa = 8.74VTVFCFAVRR212 pKa = 11.84DD213 pKa = 3.71LRR215 pKa = 11.84TSYY218 pKa = 11.27NILTPYY224 pKa = 7.03TTLDD228 pKa = 3.93LAPGTSPNLPILPAASNAVLEE249 pKa = 4.41QAKK252 pKa = 8.04QTSGNVVQDD261 pKa = 3.82SLCVSTGFDD270 pKa = 3.59GDD272 pKa = 4.1SLNSFVAASTDD283 pKa = 3.2AQTCNWFLKK292 pKa = 10.7NPWSYY297 pKa = 11.0YY298 pKa = 7.22KK299 pKa = 10.16WRR301 pKa = 11.84MEE303 pKa = 3.81YY304 pKa = 10.43RR305 pKa = 11.84KK306 pKa = 9.94VSQVSFMLPAGQGTCTRR323 pKa = 11.84YY324 pKa = 9.68FNHH327 pKa = 6.41SFKK330 pKa = 11.23LNNKK334 pKa = 7.69WSFYY338 pKa = 8.8TEE340 pKa = 4.38WAQHH344 pKa = 5.3PAFTRR349 pKa = 11.84FYY351 pKa = 10.55LIQTEE356 pKa = 4.37TQPMSTVTTGVLPSQQGFLAYY377 pKa = 9.69PPATVGLLVEE387 pKa = 4.74SKK389 pKa = 9.56TVIQRR394 pKa = 11.84VSGVTRR400 pKa = 11.84ASHH403 pKa = 5.97LRR405 pKa = 11.84EE406 pKa = 4.08PLVDD410 pKa = 3.61PTPITGNASTGAADD424 pKa = 5.37FIAPASLVSGALNMASEE441 pKa = 4.48EE442 pKa = 4.17TDD444 pKa = 3.11QKK446 pKa = 10.32QAGVFFF452 pKa = 5.24
Molecular weight: 49.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
769 |
317 |
452 |
384.5 |
43.61 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.982 ± 1.0 | 1.43 ± 0.497 |
5.202 ± 1.916 | 4.551 ± 2.332 |
5.072 ± 0.821 | 5.202 ± 1.307 |
2.081 ± 0.887 | 4.941 ± 1.277 |
7.022 ± 2.164 | 7.282 ± 0.42 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.341 ± 0.117 | 7.022 ± 0.052 |
5.202 ± 0.501 | 5.332 ± 0.988 |
4.551 ± 0.086 | 7.672 ± 1.475 |
7.932 ± 2.246 | 5.982 ± 1.201 |
2.081 ± 0.484 | 3.121 ± 0.424 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
