
Streptococcus satellite phage Javan437
Taxonomy: Viruses; unclassified bacterial viruses
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
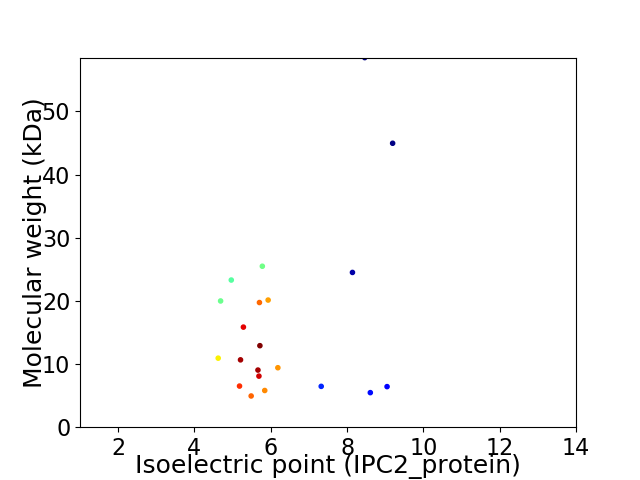
Virtual 2D-PAGE plot for 21 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZTE1|A0A4D5ZTE1_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan437 OX=2558704 GN=JavanS437_0019 PE=4 SV=1
MM1 pKa = 7.54IADD4 pKa = 3.78KK5 pKa = 11.39LEE7 pKa = 4.85AISTNLEE14 pKa = 4.11EE15 pKa = 4.13IQEE18 pKa = 4.13ILVGNGTALAKK29 pKa = 10.45RR30 pKa = 11.84EE31 pKa = 3.86ISKK34 pKa = 10.38VLNDD38 pKa = 4.47LDD40 pKa = 5.59DD41 pKa = 4.4IYY43 pKa = 11.79NEE45 pKa = 4.13LTSTEE50 pKa = 3.99YY51 pKa = 10.82HH52 pKa = 5.9EE53 pKa = 4.26QQEE56 pKa = 4.54TLKK59 pKa = 10.72EE60 pKa = 3.7QTEE63 pKa = 4.21RR64 pKa = 11.84MKK66 pKa = 10.83QRR68 pKa = 11.84VVSEE72 pKa = 4.31VIGEE76 pKa = 4.27LEE78 pKa = 3.68WRR80 pKa = 11.84EE81 pKa = 3.45YY82 pKa = 11.05SYY84 pKa = 11.97YY85 pKa = 10.94KK86 pKa = 10.66NFWGDD91 pKa = 3.24MDD93 pKa = 5.26LLKK96 pKa = 10.88CLSNFDD102 pKa = 3.56GFLFISTYY110 pKa = 10.01LAEE113 pKa = 4.1ITKK116 pKa = 10.6EE117 pKa = 3.51NDD119 pKa = 2.93YY120 pKa = 11.52PMDD123 pKa = 4.03KK124 pKa = 10.74NQVLNYY130 pKa = 9.47VWEE133 pKa = 4.22LLAIDD138 pKa = 3.6IAKK141 pKa = 10.3KK142 pKa = 10.58KK143 pKa = 9.91RR144 pKa = 11.84DD145 pKa = 3.56KK146 pKa = 11.32KK147 pKa = 10.92NLLGLWTSSIEE158 pKa = 4.07YY159 pKa = 8.46TRR161 pKa = 11.84GMMIYY166 pKa = 9.87EE167 pKa = 4.03EE168 pKa = 4.39HH169 pKa = 6.67
MM1 pKa = 7.54IADD4 pKa = 3.78KK5 pKa = 11.39LEE7 pKa = 4.85AISTNLEE14 pKa = 4.11EE15 pKa = 4.13IQEE18 pKa = 4.13ILVGNGTALAKK29 pKa = 10.45RR30 pKa = 11.84EE31 pKa = 3.86ISKK34 pKa = 10.38VLNDD38 pKa = 4.47LDD40 pKa = 5.59DD41 pKa = 4.4IYY43 pKa = 11.79NEE45 pKa = 4.13LTSTEE50 pKa = 3.99YY51 pKa = 10.82HH52 pKa = 5.9EE53 pKa = 4.26QQEE56 pKa = 4.54TLKK59 pKa = 10.72EE60 pKa = 3.7QTEE63 pKa = 4.21RR64 pKa = 11.84MKK66 pKa = 10.83QRR68 pKa = 11.84VVSEE72 pKa = 4.31VIGEE76 pKa = 4.27LEE78 pKa = 3.68WRR80 pKa = 11.84EE81 pKa = 3.45YY82 pKa = 11.05SYY84 pKa = 11.97YY85 pKa = 10.94KK86 pKa = 10.66NFWGDD91 pKa = 3.24MDD93 pKa = 5.26LLKK96 pKa = 10.88CLSNFDD102 pKa = 3.56GFLFISTYY110 pKa = 10.01LAEE113 pKa = 4.1ITKK116 pKa = 10.6EE117 pKa = 3.51NDD119 pKa = 2.93YY120 pKa = 11.52PMDD123 pKa = 4.03KK124 pKa = 10.74NQVLNYY130 pKa = 9.47VWEE133 pKa = 4.22LLAIDD138 pKa = 3.6IAKK141 pKa = 10.3KK142 pKa = 10.58KK143 pKa = 9.91RR144 pKa = 11.84DD145 pKa = 3.56KK146 pKa = 11.32KK147 pKa = 10.92NLLGLWTSSIEE158 pKa = 4.07YY159 pKa = 8.46TRR161 pKa = 11.84GMMIYY166 pKa = 9.87EE167 pKa = 4.03EE168 pKa = 4.39HH169 pKa = 6.67
Molecular weight: 19.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZNL6|A0A4D5ZNL6_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan437 OX=2558704 GN=JavanS437_0011 PE=4 SV=1
MM1 pKa = 7.87KK2 pKa = 9.27ITEE5 pKa = 4.15YY6 pKa = 10.16TKK8 pKa = 10.79KK9 pKa = 10.58DD10 pKa = 2.93GSTVYY15 pKa = 10.37RR16 pKa = 11.84SSVYY20 pKa = 10.68LGIDD24 pKa = 3.15TVTGKK29 pKa = 10.24KK30 pKa = 10.27VKK32 pKa = 8.07TTISGRR38 pKa = 11.84TKK40 pKa = 10.61RR41 pKa = 11.84EE42 pKa = 3.76LKK44 pKa = 10.7AKK46 pKa = 10.16ALQAQIDD53 pKa = 4.0FEE55 pKa = 5.43KK56 pKa = 11.01DD57 pKa = 2.64GSTVYY62 pKa = 10.61KK63 pKa = 10.4AVEE66 pKa = 3.74IKK68 pKa = 9.8TYY70 pKa = 11.23AEE72 pKa = 4.37LVEE75 pKa = 4.62NWLEE79 pKa = 4.57TYY81 pKa = 10.53CHH83 pKa = 5.45TVKK86 pKa = 10.81KK87 pKa = 9.8ATLTIIKK94 pKa = 10.1SRR96 pKa = 11.84LKK98 pKa = 10.87NYY100 pKa = 9.35LLPAFGDD107 pKa = 3.76YY108 pKa = 10.88KK109 pKa = 10.73LDD111 pKa = 3.6KK112 pKa = 9.52LTPPVIQKK120 pKa = 9.14QVNQWAKK127 pKa = 10.6EE128 pKa = 3.84YY129 pKa = 9.72NQLGKK134 pKa = 10.69GYY136 pKa = 9.89QEE138 pKa = 3.99YY139 pKa = 9.14PQLNSLNKK147 pKa = 10.15RR148 pKa = 11.84ILKK151 pKa = 9.76YY152 pKa = 10.38AVSLQVIPFNPARR165 pKa = 11.84DD166 pKa = 3.53IIVPRR171 pKa = 11.84RR172 pKa = 11.84KK173 pKa = 10.05AKK175 pKa = 10.24EE176 pKa = 4.02GQKK179 pKa = 10.55LKK181 pKa = 11.17YY182 pKa = 10.43LDD184 pKa = 4.8DD185 pKa = 5.74DD186 pKa = 4.04NLKK189 pKa = 10.78KK190 pKa = 10.49FLNYY194 pKa = 10.06LDD196 pKa = 5.07RR197 pKa = 11.84LPNTYY202 pKa = 10.78KK203 pKa = 10.76NFYY206 pKa = 9.48DD207 pKa = 3.51TVLYY211 pKa = 8.15KK212 pKa = 10.45TLLATGLRR220 pKa = 11.84IRR222 pKa = 11.84EE223 pKa = 4.08CLALKK228 pKa = 10.05WSDD231 pKa = 3.08IDD233 pKa = 4.02LQNGTLDD240 pKa = 3.42VNKK243 pKa = 9.41TLNCEE248 pKa = 4.24KK249 pKa = 10.58EE250 pKa = 4.16VTTPKK255 pKa = 9.67TKK257 pKa = 10.55SSVRR261 pKa = 11.84MIDD264 pKa = 3.65LDD266 pKa = 3.84NKK268 pKa = 8.1TVLMLRR274 pKa = 11.84LYY276 pKa = 10.7KK277 pKa = 10.34NRR279 pKa = 11.84QAQVGRR285 pKa = 11.84EE286 pKa = 3.43IGLTYY291 pKa = 10.66KK292 pKa = 10.49KK293 pKa = 10.61VFSNSFDD300 pKa = 3.44EE301 pKa = 4.33YY302 pKa = 10.78RR303 pKa = 11.84DD304 pKa = 3.44ARR306 pKa = 11.84ALRR309 pKa = 11.84SRR311 pKa = 11.84LEE313 pKa = 3.75KK314 pKa = 10.29HH315 pKa = 6.29LKK317 pKa = 10.4LSEE320 pKa = 4.09CPRR323 pKa = 11.84LTFHH327 pKa = 7.91AFRR330 pKa = 11.84HH331 pKa = 4.54THH333 pKa = 6.71ASILLNAGLPYY344 pKa = 10.43KK345 pKa = 10.2EE346 pKa = 3.68IQTRR350 pKa = 11.84LGHH353 pKa = 5.05SQISITMDD361 pKa = 3.56TYY363 pKa = 11.78SHH365 pKa = 7.28LSKK368 pKa = 10.85DD369 pKa = 3.51NKK371 pKa = 10.49KK372 pKa = 10.17NATSFYY378 pKa = 9.85EE379 pKa = 3.99KK380 pKa = 10.33AIEE383 pKa = 4.04KK384 pKa = 10.35LKK386 pKa = 11.16SSS388 pKa = 3.82
MM1 pKa = 7.87KK2 pKa = 9.27ITEE5 pKa = 4.15YY6 pKa = 10.16TKK8 pKa = 10.79KK9 pKa = 10.58DD10 pKa = 2.93GSTVYY15 pKa = 10.37RR16 pKa = 11.84SSVYY20 pKa = 10.68LGIDD24 pKa = 3.15TVTGKK29 pKa = 10.24KK30 pKa = 10.27VKK32 pKa = 8.07TTISGRR38 pKa = 11.84TKK40 pKa = 10.61RR41 pKa = 11.84EE42 pKa = 3.76LKK44 pKa = 10.7AKK46 pKa = 10.16ALQAQIDD53 pKa = 4.0FEE55 pKa = 5.43KK56 pKa = 11.01DD57 pKa = 2.64GSTVYY62 pKa = 10.61KK63 pKa = 10.4AVEE66 pKa = 3.74IKK68 pKa = 9.8TYY70 pKa = 11.23AEE72 pKa = 4.37LVEE75 pKa = 4.62NWLEE79 pKa = 4.57TYY81 pKa = 10.53CHH83 pKa = 5.45TVKK86 pKa = 10.81KK87 pKa = 9.8ATLTIIKK94 pKa = 10.1SRR96 pKa = 11.84LKK98 pKa = 10.87NYY100 pKa = 9.35LLPAFGDD107 pKa = 3.76YY108 pKa = 10.88KK109 pKa = 10.73LDD111 pKa = 3.6KK112 pKa = 9.52LTPPVIQKK120 pKa = 9.14QVNQWAKK127 pKa = 10.6EE128 pKa = 3.84YY129 pKa = 9.72NQLGKK134 pKa = 10.69GYY136 pKa = 9.89QEE138 pKa = 3.99YY139 pKa = 9.14PQLNSLNKK147 pKa = 10.15RR148 pKa = 11.84ILKK151 pKa = 9.76YY152 pKa = 10.38AVSLQVIPFNPARR165 pKa = 11.84DD166 pKa = 3.53IIVPRR171 pKa = 11.84RR172 pKa = 11.84KK173 pKa = 10.05AKK175 pKa = 10.24EE176 pKa = 4.02GQKK179 pKa = 10.55LKK181 pKa = 11.17YY182 pKa = 10.43LDD184 pKa = 4.8DD185 pKa = 5.74DD186 pKa = 4.04NLKK189 pKa = 10.78KK190 pKa = 10.49FLNYY194 pKa = 10.06LDD196 pKa = 5.07RR197 pKa = 11.84LPNTYY202 pKa = 10.78KK203 pKa = 10.76NFYY206 pKa = 9.48DD207 pKa = 3.51TVLYY211 pKa = 8.15KK212 pKa = 10.45TLLATGLRR220 pKa = 11.84IRR222 pKa = 11.84EE223 pKa = 4.08CLALKK228 pKa = 10.05WSDD231 pKa = 3.08IDD233 pKa = 4.02LQNGTLDD240 pKa = 3.42VNKK243 pKa = 9.41TLNCEE248 pKa = 4.24KK249 pKa = 10.58EE250 pKa = 4.16VTTPKK255 pKa = 9.67TKK257 pKa = 10.55SSVRR261 pKa = 11.84MIDD264 pKa = 3.65LDD266 pKa = 3.84NKK268 pKa = 8.1TVLMLRR274 pKa = 11.84LYY276 pKa = 10.7KK277 pKa = 10.34NRR279 pKa = 11.84QAQVGRR285 pKa = 11.84EE286 pKa = 3.43IGLTYY291 pKa = 10.66KK292 pKa = 10.49KK293 pKa = 10.61VFSNSFDD300 pKa = 3.44EE301 pKa = 4.33YY302 pKa = 10.78RR303 pKa = 11.84DD304 pKa = 3.44ARR306 pKa = 11.84ALRR309 pKa = 11.84SRR311 pKa = 11.84LEE313 pKa = 3.75KK314 pKa = 10.29HH315 pKa = 6.29LKK317 pKa = 10.4LSEE320 pKa = 4.09CPRR323 pKa = 11.84LTFHH327 pKa = 7.91AFRR330 pKa = 11.84HH331 pKa = 4.54THH333 pKa = 6.71ASILLNAGLPYY344 pKa = 10.43KK345 pKa = 10.2EE346 pKa = 3.68IQTRR350 pKa = 11.84LGHH353 pKa = 5.05SQISITMDD361 pKa = 3.56TYY363 pKa = 11.78SHH365 pKa = 7.28LSKK368 pKa = 10.85DD369 pKa = 3.51NKK371 pKa = 10.49KK372 pKa = 10.17NATSFYY378 pKa = 9.85EE379 pKa = 3.99KK380 pKa = 10.33AIEE383 pKa = 4.04KK384 pKa = 10.35LKK386 pKa = 11.16SSS388 pKa = 3.82
Molecular weight: 44.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3008 |
41 |
507 |
143.2 |
16.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.652 ± 0.407 | 0.465 ± 0.127 |
5.751 ± 0.36 | 9.176 ± 0.748 |
3.956 ± 0.438 | 4.82 ± 0.35 |
1.563 ± 0.294 | 7.148 ± 0.492 |
9.608 ± 0.622 | 10.173 ± 0.708 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.427 ± 0.386 | 4.754 ± 0.34 |
2.726 ± 0.434 | 4.555 ± 0.331 |
5.386 ± 0.295 | 5.618 ± 0.212 |
5.918 ± 0.416 | 4.787 ± 0.418 |
0.931 ± 0.173 | 4.588 ± 0.256 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
