
Bradymonadales bacterium V1718
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Bradymonadales; unclassified Bradymonadales
Average proteome isoelectric point is 5.58
Get precalculated fractions of proteins
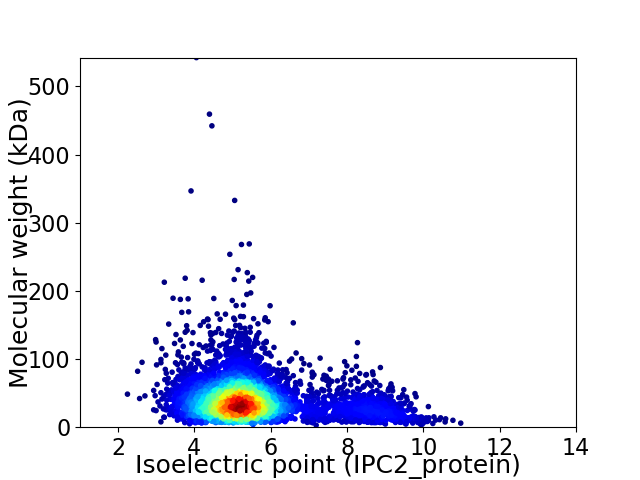
Virtual 2D-PAGE plot for 4727 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
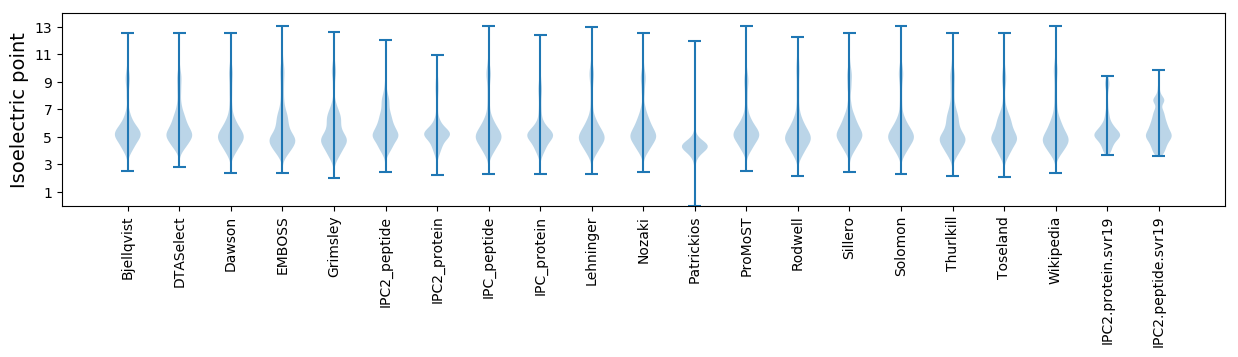
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B8XNV3|A0A5B8XNV3_9DELT Uncharacterized protein OS=Bradymonadales bacterium V1718 OX=2600177 GN=FRD01_08150 PE=4 SV=1
MM1 pKa = 7.29FRR3 pKa = 11.84LLILVALFSLGSGCSGCDD21 pKa = 3.62DD22 pKa = 4.61DD23 pKa = 6.62SGGIPNSDD31 pKa = 3.68AGNNTQDD38 pKa = 3.78MADD41 pKa = 3.85DD42 pKa = 4.61ANNLGDD48 pKa = 4.6GGQSDD53 pKa = 3.83MGEE56 pKa = 4.17PDD58 pKa = 3.91GGPSDD63 pKa = 3.9MGEE66 pKa = 4.15PDD68 pKa = 4.9ANTCSTVEE76 pKa = 4.27CNGVCCGEE84 pKa = 4.11GQEE87 pKa = 4.99CFQDD91 pKa = 4.2LCLDD95 pKa = 4.06PCAGTRR101 pKa = 11.84CGAGFEE107 pKa = 4.51LCCAEE112 pKa = 5.31ADD114 pKa = 3.37ICLGQACLTPGADD127 pKa = 3.27CTVTEE132 pKa = 4.3EE133 pKa = 4.42CAVEE137 pKa = 4.69EE138 pKa = 4.1ICEE141 pKa = 4.17PTVGKK146 pKa = 9.94CVPRR150 pKa = 11.84DD151 pKa = 3.37AVEE154 pKa = 3.94VCEE157 pKa = 5.07FIPPIGPFEE166 pKa = 4.34PEE168 pKa = 4.43VGCFWPNPAPTVNPDD183 pKa = 3.13SVHH186 pKa = 5.96VVVAPIVGNLTDD198 pKa = 4.83DD199 pKa = 4.19NGDD202 pKa = 3.61GLTNTDD208 pKa = 4.33DD209 pKa = 3.51VPEE212 pKa = 4.2IVFLTRR218 pKa = 11.84TSGCCDD224 pKa = 2.54KK225 pKa = 11.05RR226 pKa = 11.84GTLRR230 pKa = 11.84IVDD233 pKa = 4.14GRR235 pKa = 11.84CGANGEE241 pKa = 4.3MTTIASLDD249 pKa = 3.94SVVMTNDD256 pKa = 3.23AAPALGDD263 pKa = 3.82LDD265 pKa = 4.4GDD267 pKa = 4.1GVPEE271 pKa = 4.04IVAVKK276 pKa = 10.01GQNLQTNQKK285 pKa = 8.15VTPQGLVAWKK295 pKa = 10.2RR296 pKa = 11.84VTPDD300 pKa = 2.87GATWEE305 pKa = 4.28PMWEE309 pKa = 3.9NDD311 pKa = 3.24EE312 pKa = 4.36YY313 pKa = 10.6PTFGVHH319 pKa = 5.27TRR321 pKa = 11.84GGPTVGIADD330 pKa = 4.75LDD332 pKa = 4.05GDD334 pKa = 4.25GNPEE338 pKa = 3.87VFVGNVVLNGQDD350 pKa = 5.37GNLKK354 pKa = 9.34WDD356 pKa = 3.7GVANSQTPVGIGNNAFLGPSSIAADD381 pKa = 3.33VDD383 pKa = 4.11LDD385 pKa = 3.84GMQEE389 pKa = 3.91VMAGNTLYY397 pKa = 11.1SHH399 pKa = 7.7DD400 pKa = 3.59GTVRR404 pKa = 11.84WTYY407 pKa = 10.95EE408 pKa = 3.83YY409 pKa = 7.39TTSNSTCQGGLPCDD423 pKa = 4.25GYY425 pKa = 9.34TAVAEE430 pKa = 4.09FDD432 pKa = 3.8GDD434 pKa = 3.95PEE436 pKa = 4.47GEE438 pKa = 3.93IVIIRR443 pKa = 11.84LGEE446 pKa = 4.23VFVLNHH452 pKa = 7.19DD453 pKa = 3.95GTLFWQQQIVKK464 pKa = 9.92DD465 pKa = 3.55DD466 pKa = 3.64CTRR469 pKa = 11.84NEE471 pKa = 4.39SGPPTIADD479 pKa = 3.35FDD481 pKa = 4.68GDD483 pKa = 3.82GRR485 pKa = 11.84PEE487 pKa = 4.1IGTAAADD494 pKa = 4.85FYY496 pKa = 11.32TVLDD500 pKa = 4.83LDD502 pKa = 4.91CDD504 pKa = 3.74TDD506 pKa = 3.12TWEE509 pKa = 5.97DD510 pKa = 3.53DD511 pKa = 3.56GCFARR516 pKa = 11.84GVLWATPNQDD526 pKa = 2.77CSSRR530 pKa = 11.84VTASSVFDD538 pKa = 3.8FEE540 pKa = 5.69GDD542 pKa = 3.31GKK544 pKa = 11.16AEE546 pKa = 3.76MVYY549 pKa = 10.93ADD551 pKa = 3.99EE552 pKa = 4.25QNFRR556 pKa = 11.84IFDD559 pKa = 3.9GTTGAILFDD568 pKa = 4.82DD569 pKa = 5.31PSHH572 pKa = 6.98SSNTRR577 pKa = 11.84IEE579 pKa = 4.18MPIVADD585 pKa = 3.51VDD587 pKa = 3.47NDD589 pKa = 3.89GNSEE593 pKa = 4.07IVVPSATAQSIKK605 pKa = 10.46VFKK608 pKa = 10.57DD609 pKa = 3.49PSDD612 pKa = 2.9NWVRR616 pKa = 11.84TRR618 pKa = 11.84RR619 pKa = 11.84IWNQHH624 pKa = 4.85AYY626 pKa = 10.15AVTNINEE633 pKa = 4.82DD634 pKa = 3.56GTVPAQPDD642 pKa = 3.88INWLNGRR649 pKa = 11.84LNNFRR654 pKa = 11.84QNIQPGGIFDD664 pKa = 4.5APNLLIEE671 pKa = 4.51SVDD674 pKa = 3.65VRR676 pKa = 11.84GLGCGEE682 pKa = 4.16DD683 pKa = 3.53SEE685 pKa = 5.3VIIRR689 pKa = 11.84VTVSNAGALGVAPGNTLIRR708 pKa = 11.84IYY710 pKa = 10.58GSSGGDD716 pKa = 3.16TIVIDD721 pKa = 4.09DD722 pKa = 3.94TTLTTRR728 pKa = 11.84LLPGQRR734 pKa = 11.84EE735 pKa = 4.42VVEE738 pKa = 3.98VTYY741 pKa = 10.6AVPADD746 pKa = 3.28WVTNGFEE753 pKa = 3.75IGAIIDD759 pKa = 3.57PDD761 pKa = 3.5ATINEE766 pKa = 4.48CNEE769 pKa = 4.32DD770 pKa = 3.99DD771 pKa = 4.03NQGSFDD777 pKa = 3.55GANVVFSAPEE787 pKa = 4.01LEE789 pKa = 4.58VTTLTATGTTCGLTLQMPIAFTVRR813 pKa = 11.84NSGTDD818 pKa = 3.28TVPANVPLVVTGTFQGTTVEE838 pKa = 4.23IARR841 pKa = 11.84VTTSAEE847 pKa = 3.87LLVGEE852 pKa = 4.86EE853 pKa = 4.17EE854 pKa = 4.79DD855 pKa = 5.22FNLVWTVDD863 pKa = 3.7PGAPAQDD870 pKa = 3.34VTITVTVDD878 pKa = 3.14PDD880 pKa = 3.71KK881 pKa = 11.15EE882 pKa = 4.67VYY884 pKa = 10.52DD885 pKa = 4.36CDD887 pKa = 3.67EE888 pKa = 4.18QEE890 pKa = 4.21ALSVVEE896 pKa = 4.1NCRR899 pKa = 11.84ISGG902 pKa = 3.5
MM1 pKa = 7.29FRR3 pKa = 11.84LLILVALFSLGSGCSGCDD21 pKa = 3.62DD22 pKa = 4.61DD23 pKa = 6.62SGGIPNSDD31 pKa = 3.68AGNNTQDD38 pKa = 3.78MADD41 pKa = 3.85DD42 pKa = 4.61ANNLGDD48 pKa = 4.6GGQSDD53 pKa = 3.83MGEE56 pKa = 4.17PDD58 pKa = 3.91GGPSDD63 pKa = 3.9MGEE66 pKa = 4.15PDD68 pKa = 4.9ANTCSTVEE76 pKa = 4.27CNGVCCGEE84 pKa = 4.11GQEE87 pKa = 4.99CFQDD91 pKa = 4.2LCLDD95 pKa = 4.06PCAGTRR101 pKa = 11.84CGAGFEE107 pKa = 4.51LCCAEE112 pKa = 5.31ADD114 pKa = 3.37ICLGQACLTPGADD127 pKa = 3.27CTVTEE132 pKa = 4.3EE133 pKa = 4.42CAVEE137 pKa = 4.69EE138 pKa = 4.1ICEE141 pKa = 4.17PTVGKK146 pKa = 9.94CVPRR150 pKa = 11.84DD151 pKa = 3.37AVEE154 pKa = 3.94VCEE157 pKa = 5.07FIPPIGPFEE166 pKa = 4.34PEE168 pKa = 4.43VGCFWPNPAPTVNPDD183 pKa = 3.13SVHH186 pKa = 5.96VVVAPIVGNLTDD198 pKa = 4.83DD199 pKa = 4.19NGDD202 pKa = 3.61GLTNTDD208 pKa = 4.33DD209 pKa = 3.51VPEE212 pKa = 4.2IVFLTRR218 pKa = 11.84TSGCCDD224 pKa = 2.54KK225 pKa = 11.05RR226 pKa = 11.84GTLRR230 pKa = 11.84IVDD233 pKa = 4.14GRR235 pKa = 11.84CGANGEE241 pKa = 4.3MTTIASLDD249 pKa = 3.94SVVMTNDD256 pKa = 3.23AAPALGDD263 pKa = 3.82LDD265 pKa = 4.4GDD267 pKa = 4.1GVPEE271 pKa = 4.04IVAVKK276 pKa = 10.01GQNLQTNQKK285 pKa = 8.15VTPQGLVAWKK295 pKa = 10.2RR296 pKa = 11.84VTPDD300 pKa = 2.87GATWEE305 pKa = 4.28PMWEE309 pKa = 3.9NDD311 pKa = 3.24EE312 pKa = 4.36YY313 pKa = 10.6PTFGVHH319 pKa = 5.27TRR321 pKa = 11.84GGPTVGIADD330 pKa = 4.75LDD332 pKa = 4.05GDD334 pKa = 4.25GNPEE338 pKa = 3.87VFVGNVVLNGQDD350 pKa = 5.37GNLKK354 pKa = 9.34WDD356 pKa = 3.7GVANSQTPVGIGNNAFLGPSSIAADD381 pKa = 3.33VDD383 pKa = 4.11LDD385 pKa = 3.84GMQEE389 pKa = 3.91VMAGNTLYY397 pKa = 11.1SHH399 pKa = 7.7DD400 pKa = 3.59GTVRR404 pKa = 11.84WTYY407 pKa = 10.95EE408 pKa = 3.83YY409 pKa = 7.39TTSNSTCQGGLPCDD423 pKa = 4.25GYY425 pKa = 9.34TAVAEE430 pKa = 4.09FDD432 pKa = 3.8GDD434 pKa = 3.95PEE436 pKa = 4.47GEE438 pKa = 3.93IVIIRR443 pKa = 11.84LGEE446 pKa = 4.23VFVLNHH452 pKa = 7.19DD453 pKa = 3.95GTLFWQQQIVKK464 pKa = 9.92DD465 pKa = 3.55DD466 pKa = 3.64CTRR469 pKa = 11.84NEE471 pKa = 4.39SGPPTIADD479 pKa = 3.35FDD481 pKa = 4.68GDD483 pKa = 3.82GRR485 pKa = 11.84PEE487 pKa = 4.1IGTAAADD494 pKa = 4.85FYY496 pKa = 11.32TVLDD500 pKa = 4.83LDD502 pKa = 4.91CDD504 pKa = 3.74TDD506 pKa = 3.12TWEE509 pKa = 5.97DD510 pKa = 3.53DD511 pKa = 3.56GCFARR516 pKa = 11.84GVLWATPNQDD526 pKa = 2.77CSSRR530 pKa = 11.84VTASSVFDD538 pKa = 3.8FEE540 pKa = 5.69GDD542 pKa = 3.31GKK544 pKa = 11.16AEE546 pKa = 3.76MVYY549 pKa = 10.93ADD551 pKa = 3.99EE552 pKa = 4.25QNFRR556 pKa = 11.84IFDD559 pKa = 3.9GTTGAILFDD568 pKa = 4.82DD569 pKa = 5.31PSHH572 pKa = 6.98SSNTRR577 pKa = 11.84IEE579 pKa = 4.18MPIVADD585 pKa = 3.51VDD587 pKa = 3.47NDD589 pKa = 3.89GNSEE593 pKa = 4.07IVVPSATAQSIKK605 pKa = 10.46VFKK608 pKa = 10.57DD609 pKa = 3.49PSDD612 pKa = 2.9NWVRR616 pKa = 11.84TRR618 pKa = 11.84RR619 pKa = 11.84IWNQHH624 pKa = 4.85AYY626 pKa = 10.15AVTNINEE633 pKa = 4.82DD634 pKa = 3.56GTVPAQPDD642 pKa = 3.88INWLNGRR649 pKa = 11.84LNNFRR654 pKa = 11.84QNIQPGGIFDD664 pKa = 4.5APNLLIEE671 pKa = 4.51SVDD674 pKa = 3.65VRR676 pKa = 11.84GLGCGEE682 pKa = 4.16DD683 pKa = 3.53SEE685 pKa = 5.3VIIRR689 pKa = 11.84VTVSNAGALGVAPGNTLIRR708 pKa = 11.84IYY710 pKa = 10.58GSSGGDD716 pKa = 3.16TIVIDD721 pKa = 4.09DD722 pKa = 3.94TTLTTRR728 pKa = 11.84LLPGQRR734 pKa = 11.84EE735 pKa = 4.42VVEE738 pKa = 3.98VTYY741 pKa = 10.6AVPADD746 pKa = 3.28WVTNGFEE753 pKa = 3.75IGAIIDD759 pKa = 3.57PDD761 pKa = 3.5ATINEE766 pKa = 4.48CNEE769 pKa = 4.32DD770 pKa = 3.99DD771 pKa = 4.03NQGSFDD777 pKa = 3.55GANVVFSAPEE787 pKa = 4.01LEE789 pKa = 4.58VTTLTATGTTCGLTLQMPIAFTVRR813 pKa = 11.84NSGTDD818 pKa = 3.28TVPANVPLVVTGTFQGTTVEE838 pKa = 4.23IARR841 pKa = 11.84VTTSAEE847 pKa = 3.87LLVGEE852 pKa = 4.86EE853 pKa = 4.17EE854 pKa = 4.79DD855 pKa = 5.22FNLVWTVDD863 pKa = 3.7PGAPAQDD870 pKa = 3.34VTITVTVDD878 pKa = 3.14PDD880 pKa = 3.71KK881 pKa = 11.15EE882 pKa = 4.67VYY884 pKa = 10.52DD885 pKa = 4.36CDD887 pKa = 3.67EE888 pKa = 4.18QEE890 pKa = 4.21ALSVVEE896 pKa = 4.1NCRR899 pKa = 11.84ISGG902 pKa = 3.5
Molecular weight: 95.44 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B8XS27|A0A5B8XS27_9DELT MBL fold metallo-hydrolase OS=Bradymonadales bacterium V1718 OX=2600177 GN=FRD01_10675 PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.24QPSKK10 pKa = 7.88VKK12 pKa = 10.25RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84THH17 pKa = 5.65GFRR20 pKa = 11.84ARR22 pKa = 11.84MADD25 pKa = 2.99AGGRR29 pKa = 11.84KK30 pKa = 8.93VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 7.87RR42 pKa = 11.84LAVQRR47 pKa = 11.84HH48 pKa = 4.75KK49 pKa = 10.93KK50 pKa = 9.57
MM1 pKa = 7.74SKK3 pKa = 8.95RR4 pKa = 11.84TYY6 pKa = 10.24QPSKK10 pKa = 7.88VKK12 pKa = 10.25RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84THH17 pKa = 5.65GFRR20 pKa = 11.84ARR22 pKa = 11.84MADD25 pKa = 2.99AGGRR29 pKa = 11.84KK30 pKa = 8.93VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 7.87RR42 pKa = 11.84LAVQRR47 pKa = 11.84HH48 pKa = 4.75KK49 pKa = 10.93KK50 pKa = 9.57
Molecular weight: 5.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1827870 |
24 |
5142 |
386.7 |
42.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.412 ± 0.032 | 1.347 ± 0.039 |
6.196 ± 0.04 | 7.604 ± 0.039 |
4.072 ± 0.02 | 7.843 ± 0.04 |
1.989 ± 0.019 | 4.968 ± 0.023 |
3.912 ± 0.035 | 9.821 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.22 ± 0.017 | 3.263 ± 0.026 |
4.946 ± 0.026 | 3.41 ± 0.018 |
5.955 ± 0.037 | 6.344 ± 0.027 |
5.37 ± 0.03 | 7.537 ± 0.029 |
1.405 ± 0.017 | 2.386 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
