
Flavobacteriaceae bacterium 14752
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; unclassified Flavobacteriaceae
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
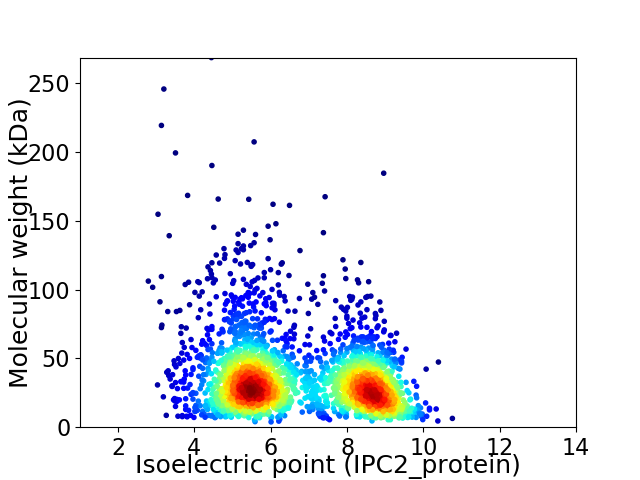
Virtual 2D-PAGE plot for 2559 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
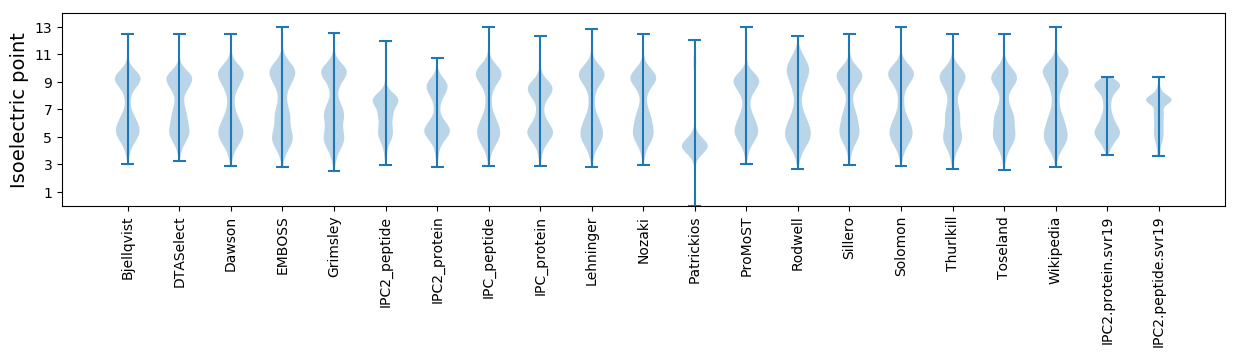
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A426KL53|A0A426KL53_9FLAO ATP-dependent 6-phosphofructokinase OS=Flavobacteriaceae bacterium 14752 OX=2491711 GN=pfkA PE=3 SV=1
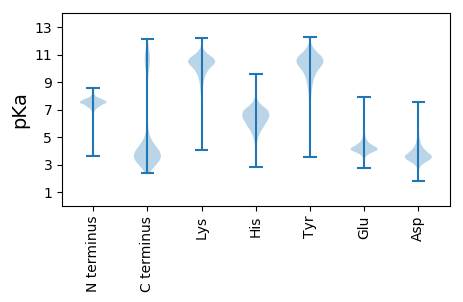
MM1 pKa = 7.8KK2 pKa = 10.38YY3 pKa = 10.45LYY5 pKa = 10.76LAFLSILFIQFSFAQTATSNDD26 pKa = 3.53SLTLNEE32 pKa = 4.03VLEE35 pKa = 4.51FMVEE39 pKa = 4.18DD40 pKa = 4.9LDD42 pKa = 4.42CASISNITSPNNAQMHH58 pKa = 5.43NQGFSSYY65 pKa = 11.42GSFQVQNEE73 pKa = 4.17PNFPFDD79 pKa = 3.35SGIVLSTTSVSDD91 pKa = 4.64LEE93 pKa = 5.23NITNNGDD100 pKa = 3.87SQWPGDD106 pKa = 3.87SDD108 pKa = 3.96LAALIQEE115 pKa = 4.88PGNTHH120 pKa = 6.28NATVIEE126 pKa = 4.14FDD128 pKa = 4.69FIPFRR133 pKa = 11.84EE134 pKa = 4.07EE135 pKa = 3.85LKK137 pKa = 10.66VDD139 pKa = 3.47YY140 pKa = 10.83LLASDD145 pKa = 4.96EE146 pKa = 4.59YY147 pKa = 10.72PVFVCDD153 pKa = 5.03FADD156 pKa = 3.57TFAFIISGPGISNVNPYY173 pKa = 10.93DD174 pKa = 3.76HH175 pKa = 7.8DD176 pKa = 5.04ANPNTPEE183 pKa = 3.92VNLDD187 pKa = 3.52LGGLNIATLPGTTIPVNPTNIHH209 pKa = 7.43DD210 pKa = 3.96MTTDD214 pKa = 3.44CTGSMGEE221 pKa = 3.96FAVPQFFDD229 pKa = 3.7AQAFDD234 pKa = 3.9NNIISFTGQTLPLTAKK250 pKa = 10.07VDD252 pKa = 5.21LIPGQTYY259 pKa = 10.36HH260 pKa = 7.26IEE262 pKa = 4.01LKK264 pKa = 10.49IADD267 pKa = 4.35RR268 pKa = 11.84GDD270 pKa = 3.5TVLNSAVFIDD280 pKa = 3.82ADD282 pKa = 3.9SFEE285 pKa = 4.78MGTIPEE291 pKa = 4.6DD292 pKa = 3.39LPYY295 pKa = 10.97EE296 pKa = 4.28PGLPVEE302 pKa = 5.87LPEE305 pKa = 4.72CWTTSDD311 pKa = 3.45TASFDD316 pKa = 3.44ILNTCSEE323 pKa = 4.12TSEE326 pKa = 5.33NYY328 pKa = 9.59LQLYY332 pKa = 8.75GGNYY336 pKa = 9.68SLQTAAVDD344 pKa = 3.67TDD346 pKa = 3.67GVAGVNISWDD356 pKa = 3.62MLNGCNDD363 pKa = 2.82IAEE366 pKa = 4.47AGKK369 pKa = 10.06NLLVEE374 pKa = 4.19YY375 pKa = 10.53FNGNDD380 pKa = 3.27WQLLADD386 pKa = 4.87IDD388 pKa = 4.86PISIPVANSNSSDD401 pKa = 2.48NWMTVNYY408 pKa = 8.51TVTSGMNKK416 pKa = 9.48NFTLRR421 pKa = 11.84FSRR424 pKa = 11.84QSGNNQQDD432 pKa = 4.23DD433 pKa = 3.57ISIANLSITEE443 pKa = 3.96QTLSNEE449 pKa = 4.13EE450 pKa = 4.08FSVDD454 pKa = 3.13TFKK457 pKa = 10.72IYY459 pKa = 10.16PNPVSDD465 pKa = 3.57ILTIEE470 pKa = 4.45TLNSTQIDD478 pKa = 3.76QIEE481 pKa = 4.58IIDD484 pKa = 3.76IQGKK488 pKa = 8.24ILKK491 pKa = 6.88TTNQLEE497 pKa = 4.11IDD499 pKa = 4.1VQSLSTGLYY508 pKa = 7.59FAKK511 pKa = 10.33ISRR514 pKa = 11.84QNSYY518 pKa = 10.19IIKK521 pKa = 10.3RR522 pKa = 11.84FIKK525 pKa = 10.4KK526 pKa = 9.89
MM1 pKa = 7.8KK2 pKa = 10.38YY3 pKa = 10.45LYY5 pKa = 10.76LAFLSILFIQFSFAQTATSNDD26 pKa = 3.53SLTLNEE32 pKa = 4.03VLEE35 pKa = 4.51FMVEE39 pKa = 4.18DD40 pKa = 4.9LDD42 pKa = 4.42CASISNITSPNNAQMHH58 pKa = 5.43NQGFSSYY65 pKa = 11.42GSFQVQNEE73 pKa = 4.17PNFPFDD79 pKa = 3.35SGIVLSTTSVSDD91 pKa = 4.64LEE93 pKa = 5.23NITNNGDD100 pKa = 3.87SQWPGDD106 pKa = 3.87SDD108 pKa = 3.96LAALIQEE115 pKa = 4.88PGNTHH120 pKa = 6.28NATVIEE126 pKa = 4.14FDD128 pKa = 4.69FIPFRR133 pKa = 11.84EE134 pKa = 4.07EE135 pKa = 3.85LKK137 pKa = 10.66VDD139 pKa = 3.47YY140 pKa = 10.83LLASDD145 pKa = 4.96EE146 pKa = 4.59YY147 pKa = 10.72PVFVCDD153 pKa = 5.03FADD156 pKa = 3.57TFAFIISGPGISNVNPYY173 pKa = 10.93DD174 pKa = 3.76HH175 pKa = 7.8DD176 pKa = 5.04ANPNTPEE183 pKa = 3.92VNLDD187 pKa = 3.52LGGLNIATLPGTTIPVNPTNIHH209 pKa = 7.43DD210 pKa = 3.96MTTDD214 pKa = 3.44CTGSMGEE221 pKa = 3.96FAVPQFFDD229 pKa = 3.7AQAFDD234 pKa = 3.9NNIISFTGQTLPLTAKK250 pKa = 10.07VDD252 pKa = 5.21LIPGQTYY259 pKa = 10.36HH260 pKa = 7.26IEE262 pKa = 4.01LKK264 pKa = 10.49IADD267 pKa = 4.35RR268 pKa = 11.84GDD270 pKa = 3.5TVLNSAVFIDD280 pKa = 3.82ADD282 pKa = 3.9SFEE285 pKa = 4.78MGTIPEE291 pKa = 4.6DD292 pKa = 3.39LPYY295 pKa = 10.97EE296 pKa = 4.28PGLPVEE302 pKa = 5.87LPEE305 pKa = 4.72CWTTSDD311 pKa = 3.45TASFDD316 pKa = 3.44ILNTCSEE323 pKa = 4.12TSEE326 pKa = 5.33NYY328 pKa = 9.59LQLYY332 pKa = 8.75GGNYY336 pKa = 9.68SLQTAAVDD344 pKa = 3.67TDD346 pKa = 3.67GVAGVNISWDD356 pKa = 3.62MLNGCNDD363 pKa = 2.82IAEE366 pKa = 4.47AGKK369 pKa = 10.06NLLVEE374 pKa = 4.19YY375 pKa = 10.53FNGNDD380 pKa = 3.27WQLLADD386 pKa = 4.87IDD388 pKa = 4.86PISIPVANSNSSDD401 pKa = 2.48NWMTVNYY408 pKa = 8.51TVTSGMNKK416 pKa = 9.48NFTLRR421 pKa = 11.84FSRR424 pKa = 11.84QSGNNQQDD432 pKa = 4.23DD433 pKa = 3.57ISIANLSITEE443 pKa = 3.96QTLSNEE449 pKa = 4.13EE450 pKa = 4.08FSVDD454 pKa = 3.13TFKK457 pKa = 10.72IYY459 pKa = 10.16PNPVSDD465 pKa = 3.57ILTIEE470 pKa = 4.45TLNSTQIDD478 pKa = 3.76QIEE481 pKa = 4.58IIDD484 pKa = 3.76IQGKK488 pKa = 8.24ILKK491 pKa = 6.88TTNQLEE497 pKa = 4.11IDD499 pKa = 4.1VQSLSTGLYY508 pKa = 7.59FAKK511 pKa = 10.33ISRR514 pKa = 11.84QNSYY518 pKa = 10.19IIKK521 pKa = 10.3RR522 pKa = 11.84FIKK525 pKa = 10.4KK526 pKa = 9.89
Molecular weight: 57.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3R8P6V5|A0A3R8P6V5_9FLAO Uncharacterized protein OS=Flavobacteriaceae bacterium 14752 OX=2491711 GN=EIG84_03770 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 8.84ILKK32 pKa = 9.62RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.12GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.62LSVSSEE47 pKa = 3.22TSMRR51 pKa = 11.84RR52 pKa = 11.84KK53 pKa = 9.82
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 10.15RR10 pKa = 11.84KK11 pKa = 9.6RR12 pKa = 11.84KK13 pKa = 8.29NKK15 pKa = 9.34HH16 pKa = 4.03GFRR19 pKa = 11.84EE20 pKa = 4.2RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 8.84ILKK32 pKa = 9.62RR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.12GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.62LSVSSEE47 pKa = 3.22TSMRR51 pKa = 11.84RR52 pKa = 11.84KK53 pKa = 9.82
Molecular weight: 6.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
856683 |
35 |
2367 |
334.8 |
38.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.891 ± 0.04 | 0.734 ± 0.015 |
5.989 ± 0.044 | 6.469 ± 0.044 |
5.515 ± 0.041 | 5.875 ± 0.049 |
1.888 ± 0.024 | 7.993 ± 0.045 |
8.239 ± 0.077 | 9.406 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.037 ± 0.022 | 6.348 ± 0.051 |
3.369 ± 0.026 | 4.133 ± 0.03 |
3.281 ± 0.033 | 6.578 ± 0.042 |
5.308 ± 0.047 | 5.952 ± 0.032 |
1.02 ± 0.018 | 3.974 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
