
Gremmeniella abietina RNA virus L1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
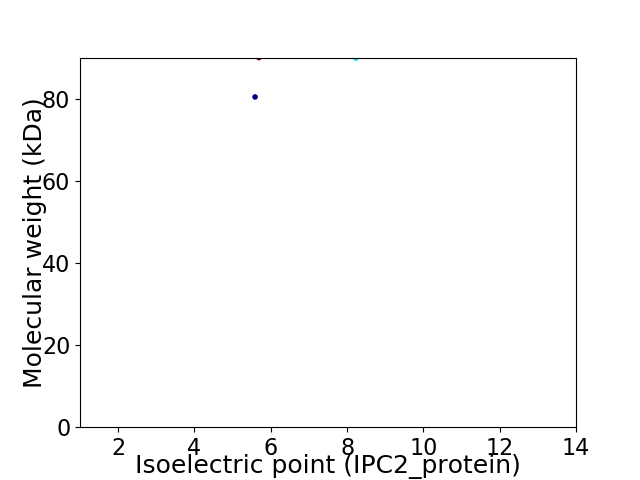
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q99AT4|Q99AT4_9VIRU Putative coat protein OS=Gremmeniella abietina RNA virus L1 OX=152217 PE=4 SV=1
MM1 pKa = 7.45ANAYY5 pKa = 7.02EE6 pKa = 4.69TNFQTANASPLTGTLADD23 pKa = 4.05PVGGTIQADD32 pKa = 3.52HH33 pKa = 6.03YY34 pKa = 9.81RR35 pKa = 11.84RR36 pKa = 11.84YY37 pKa = 9.59RR38 pKa = 11.84AGVFMSVPDD47 pKa = 3.77MGHH50 pKa = 5.47VSNVGRR56 pKa = 11.84SIFYY60 pKa = 10.17EE61 pKa = 4.15VGRR64 pKa = 11.84HH65 pKa = 5.22HH66 pKa = 7.26GRR68 pKa = 11.84ARR70 pKa = 11.84DD71 pKa = 3.8LLAAPHH77 pKa = 6.81AEE79 pKa = 5.06AIPIDD84 pKa = 3.98CSVDD88 pKa = 2.99INAAEE93 pKa = 4.02AANFEE98 pKa = 4.05GMARR102 pKa = 11.84RR103 pKa = 11.84FSNFSPQWVKK113 pKa = 10.4MDD115 pKa = 4.12LAGMVEE121 pKa = 4.37RR122 pKa = 11.84LAKK125 pKa = 10.19GVTAAGVFGGIDD137 pKa = 3.16TGTLRR142 pKa = 11.84GGQAIRR148 pKa = 11.84VTALGTLDD156 pKa = 3.78SPQTASINSVFIPRR170 pKa = 11.84TVDD173 pKa = 3.11TVGNDD178 pKa = 2.9HH179 pKa = 6.29VFAVLVSAANGEE191 pKa = 4.42GAAVTTDD198 pKa = 3.57VVRR201 pKa = 11.84LDD203 pKa = 4.14ANTNQPMIPAVAGASFATACVEE225 pKa = 3.88ALRR228 pKa = 11.84ILGANMEE235 pKa = 4.31ASGAGDD241 pKa = 3.24VFAYY245 pKa = 10.27AVTRR249 pKa = 11.84GVHH252 pKa = 5.91AGVSVVAHH260 pKa = 6.03TDD262 pKa = 2.89EE263 pKa = 5.22GGWFRR268 pKa = 11.84NVLRR272 pKa = 11.84HH273 pKa = 6.18DD274 pKa = 4.3RR275 pKa = 11.84FRR277 pKa = 11.84VPYY280 pKa = 10.4GGINQGLRR288 pKa = 11.84QYY290 pKa = 10.22PALPALASTSPSSIAAWADD309 pKa = 3.77AIALKK314 pKa = 9.18TAAAVAHH321 pKa = 6.94CDD323 pKa = 3.43PLVPGEE329 pKa = 4.32GGWYY333 pKa = 9.51PSVFTAGQNGTSAPGSSEE351 pKa = 3.86SAEE354 pKa = 4.19VGDD357 pKa = 4.29QEE359 pKa = 4.2ARR361 pKa = 11.84SIGRR365 pKa = 11.84QIASDD370 pKa = 3.27IGRR373 pKa = 11.84FAPAYY378 pKa = 9.78CRR380 pKa = 11.84ALGTLFGLHH389 pKa = 5.92TSSGVAEE396 pKa = 4.25SHH398 pKa = 6.63LSTVAMRR405 pKa = 11.84ALTQDD410 pKa = 3.36VNTDD414 pKa = 2.42RR415 pKa = 11.84HH416 pKa = 5.8LRR418 pKa = 11.84HH419 pKa = 5.81EE420 pKa = 4.46TVAPYY425 pKa = 9.78FWIEE429 pKa = 3.92PTSLIAVNAFGTPAEE444 pKa = 4.19AEE446 pKa = 4.7GYY448 pKa = 9.32GSKK451 pKa = 8.79VTPGDD456 pKa = 3.66TRR458 pKa = 11.84SEE460 pKa = 4.06PCFEE464 pKa = 4.83RR465 pKa = 11.84FRR467 pKa = 11.84LLQKK471 pKa = 10.93GSTANHH477 pKa = 4.52MTVAFKK483 pKa = 10.39MRR485 pKa = 11.84TARR488 pKa = 11.84TSALVSAYY496 pKa = 9.94AAAPAPLADD505 pKa = 3.76LKK507 pKa = 11.05LYY509 pKa = 10.7QFDD512 pKa = 3.78EE513 pKa = 4.94SSVVLPGDD521 pKa = 3.47QLPTQGDD528 pKa = 3.99VAHH531 pKa = 6.79KK532 pKa = 9.99HH533 pKa = 5.43AAADD537 pKa = 3.92PLSSYY542 pKa = 9.43LWKK545 pKa = 10.49RR546 pKa = 11.84GQSCFPAPAEE556 pKa = 4.37FINTNSNYY564 pKa = 8.64GAKK567 pKa = 9.86VRR569 pKa = 11.84LVTWDD574 pKa = 3.8DD575 pKa = 4.0DD576 pKa = 4.65FNASLSDD583 pKa = 3.74LPNDD587 pKa = 3.41EE588 pKa = 4.52EE589 pKa = 4.39MATGVVTFRR598 pKa = 11.84VTVPTGLASAGSNAEE613 pKa = 3.59NRR615 pKa = 11.84EE616 pKa = 3.94AKK618 pKa = 10.05RR619 pKa = 11.84ARR621 pKa = 11.84TRR623 pKa = 11.84GAIALAQAVLRR634 pKa = 11.84ARR636 pKa = 11.84ATGLAVSPSMEE647 pKa = 4.02ISDD650 pKa = 4.17VPPTFDD656 pKa = 4.34TPPLLPAPTYY666 pKa = 11.19SDD668 pKa = 3.52TQSASGPRR676 pKa = 11.84DD677 pKa = 3.64PGPVPVLAVGGGLDD691 pKa = 3.22VGRR694 pKa = 11.84VARR697 pKa = 11.84GAPLPPTAHH706 pKa = 6.87HH707 pKa = 6.25MPQRR711 pKa = 11.84APRR714 pKa = 11.84IAGAVVPPGGPVAGPPAATGGPAAPNVPPPPPAGPLDD751 pKa = 4.01SQDD754 pKa = 4.39PLPAPPAHH762 pKa = 6.62LPAEE766 pKa = 4.36ATAAEE771 pKa = 4.39AVPAQQ776 pKa = 3.62
MM1 pKa = 7.45ANAYY5 pKa = 7.02EE6 pKa = 4.69TNFQTANASPLTGTLADD23 pKa = 4.05PVGGTIQADD32 pKa = 3.52HH33 pKa = 6.03YY34 pKa = 9.81RR35 pKa = 11.84RR36 pKa = 11.84YY37 pKa = 9.59RR38 pKa = 11.84AGVFMSVPDD47 pKa = 3.77MGHH50 pKa = 5.47VSNVGRR56 pKa = 11.84SIFYY60 pKa = 10.17EE61 pKa = 4.15VGRR64 pKa = 11.84HH65 pKa = 5.22HH66 pKa = 7.26GRR68 pKa = 11.84ARR70 pKa = 11.84DD71 pKa = 3.8LLAAPHH77 pKa = 6.81AEE79 pKa = 5.06AIPIDD84 pKa = 3.98CSVDD88 pKa = 2.99INAAEE93 pKa = 4.02AANFEE98 pKa = 4.05GMARR102 pKa = 11.84RR103 pKa = 11.84FSNFSPQWVKK113 pKa = 10.4MDD115 pKa = 4.12LAGMVEE121 pKa = 4.37RR122 pKa = 11.84LAKK125 pKa = 10.19GVTAAGVFGGIDD137 pKa = 3.16TGTLRR142 pKa = 11.84GGQAIRR148 pKa = 11.84VTALGTLDD156 pKa = 3.78SPQTASINSVFIPRR170 pKa = 11.84TVDD173 pKa = 3.11TVGNDD178 pKa = 2.9HH179 pKa = 6.29VFAVLVSAANGEE191 pKa = 4.42GAAVTTDD198 pKa = 3.57VVRR201 pKa = 11.84LDD203 pKa = 4.14ANTNQPMIPAVAGASFATACVEE225 pKa = 3.88ALRR228 pKa = 11.84ILGANMEE235 pKa = 4.31ASGAGDD241 pKa = 3.24VFAYY245 pKa = 10.27AVTRR249 pKa = 11.84GVHH252 pKa = 5.91AGVSVVAHH260 pKa = 6.03TDD262 pKa = 2.89EE263 pKa = 5.22GGWFRR268 pKa = 11.84NVLRR272 pKa = 11.84HH273 pKa = 6.18DD274 pKa = 4.3RR275 pKa = 11.84FRR277 pKa = 11.84VPYY280 pKa = 10.4GGINQGLRR288 pKa = 11.84QYY290 pKa = 10.22PALPALASTSPSSIAAWADD309 pKa = 3.77AIALKK314 pKa = 9.18TAAAVAHH321 pKa = 6.94CDD323 pKa = 3.43PLVPGEE329 pKa = 4.32GGWYY333 pKa = 9.51PSVFTAGQNGTSAPGSSEE351 pKa = 3.86SAEE354 pKa = 4.19VGDD357 pKa = 4.29QEE359 pKa = 4.2ARR361 pKa = 11.84SIGRR365 pKa = 11.84QIASDD370 pKa = 3.27IGRR373 pKa = 11.84FAPAYY378 pKa = 9.78CRR380 pKa = 11.84ALGTLFGLHH389 pKa = 5.92TSSGVAEE396 pKa = 4.25SHH398 pKa = 6.63LSTVAMRR405 pKa = 11.84ALTQDD410 pKa = 3.36VNTDD414 pKa = 2.42RR415 pKa = 11.84HH416 pKa = 5.8LRR418 pKa = 11.84HH419 pKa = 5.81EE420 pKa = 4.46TVAPYY425 pKa = 9.78FWIEE429 pKa = 3.92PTSLIAVNAFGTPAEE444 pKa = 4.19AEE446 pKa = 4.7GYY448 pKa = 9.32GSKK451 pKa = 8.79VTPGDD456 pKa = 3.66TRR458 pKa = 11.84SEE460 pKa = 4.06PCFEE464 pKa = 4.83RR465 pKa = 11.84FRR467 pKa = 11.84LLQKK471 pKa = 10.93GSTANHH477 pKa = 4.52MTVAFKK483 pKa = 10.39MRR485 pKa = 11.84TARR488 pKa = 11.84TSALVSAYY496 pKa = 9.94AAAPAPLADD505 pKa = 3.76LKK507 pKa = 11.05LYY509 pKa = 10.7QFDD512 pKa = 3.78EE513 pKa = 4.94SSVVLPGDD521 pKa = 3.47QLPTQGDD528 pKa = 3.99VAHH531 pKa = 6.79KK532 pKa = 9.99HH533 pKa = 5.43AAADD537 pKa = 3.92PLSSYY542 pKa = 9.43LWKK545 pKa = 10.49RR546 pKa = 11.84GQSCFPAPAEE556 pKa = 4.37FINTNSNYY564 pKa = 8.64GAKK567 pKa = 9.86VRR569 pKa = 11.84LVTWDD574 pKa = 3.8DD575 pKa = 4.0DD576 pKa = 4.65FNASLSDD583 pKa = 3.74LPNDD587 pKa = 3.41EE588 pKa = 4.52EE589 pKa = 4.39MATGVVTFRR598 pKa = 11.84VTVPTGLASAGSNAEE613 pKa = 3.59NRR615 pKa = 11.84EE616 pKa = 3.94AKK618 pKa = 10.05RR619 pKa = 11.84ARR621 pKa = 11.84TRR623 pKa = 11.84GAIALAQAVLRR634 pKa = 11.84ARR636 pKa = 11.84ATGLAVSPSMEE647 pKa = 4.02ISDD650 pKa = 4.17VPPTFDD656 pKa = 4.34TPPLLPAPTYY666 pKa = 11.19SDD668 pKa = 3.52TQSASGPRR676 pKa = 11.84DD677 pKa = 3.64PGPVPVLAVGGGLDD691 pKa = 3.22VGRR694 pKa = 11.84VARR697 pKa = 11.84GAPLPPTAHH706 pKa = 6.87HH707 pKa = 6.25MPQRR711 pKa = 11.84APRR714 pKa = 11.84IAGAVVPPGGPVAGPPAATGGPAAPNVPPPPPAGPLDD751 pKa = 4.01SQDD754 pKa = 4.39PLPAPPAHH762 pKa = 6.62LPAEE766 pKa = 4.36ATAAEE771 pKa = 4.39AVPAQQ776 pKa = 3.62
Molecular weight: 80.53 kDa
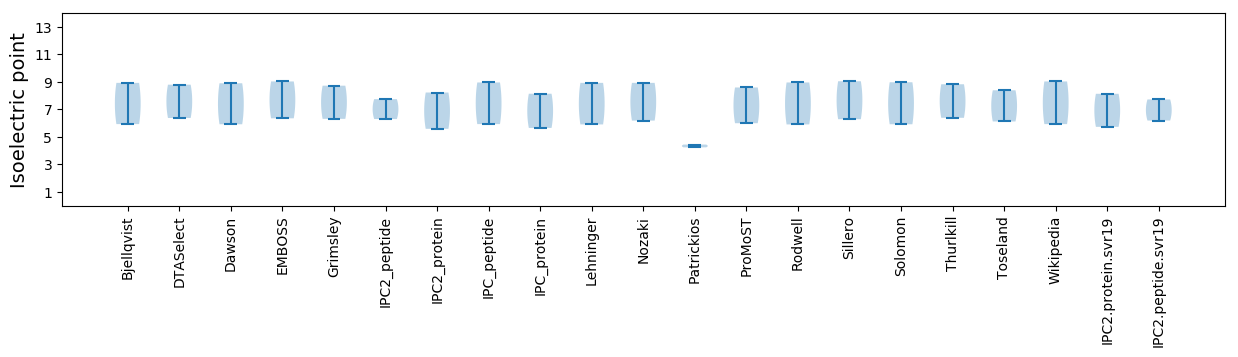
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q99AT4|Q99AT4_9VIRU Putative coat protein OS=Gremmeniella abietina RNA virus L1 OX=152217 PE=4 SV=1
MM1 pKa = 7.53IEE3 pKa = 4.23SPVSVRR9 pKa = 11.84ANEE12 pKa = 3.86AGIVGQYY19 pKa = 11.03LKK21 pKa = 11.21GLLNMEE27 pKa = 4.36WASGVMVLSFSQQISEE43 pKa = 4.43VYY45 pKa = 10.21KK46 pKa = 9.26PTFNGLRR53 pKa = 11.84PTDD56 pKa = 3.86LQRR59 pKa = 11.84AAAAYY64 pKa = 7.87LVPDD68 pKa = 4.48FPVQVRR74 pKa = 11.84IEE76 pKa = 4.17RR77 pKa = 11.84GSVLSLLSQVIDD89 pKa = 3.9PPARR93 pKa = 11.84VTDD96 pKa = 3.48RR97 pKa = 11.84CSFRR101 pKa = 11.84WLCDD105 pKa = 3.48PKK107 pKa = 11.01ASYY110 pKa = 10.66AAFPPKK116 pKa = 9.91QHH118 pKa = 6.89PGAMNKK124 pKa = 9.3VNVYY128 pKa = 9.74LNEE131 pKa = 4.12VASSLLSLDD140 pKa = 3.34PAAYY144 pKa = 10.2SSASQALWPHH154 pKa = 6.4KK155 pKa = 10.68GKK157 pKa = 10.17IANDD161 pKa = 3.31QASAIILYY169 pKa = 10.25GYY171 pKa = 8.74GLRR174 pKa = 11.84AQAVPDD180 pKa = 3.87AMHH183 pKa = 6.32VAATLATTPDD193 pKa = 3.35LAKK196 pKa = 10.91ALTNFLKK203 pKa = 10.29ATGANGSRR211 pKa = 11.84LGALLCEE218 pKa = 4.72SNVLLGRR225 pKa = 11.84AAGPADD231 pKa = 3.67LSEE234 pKa = 4.07EE235 pKa = 3.73ARR237 pKa = 11.84YY238 pKa = 7.94RR239 pKa = 11.84TSSDD243 pKa = 3.63VEE245 pKa = 3.92SHH247 pKa = 6.78LAIFSDD253 pKa = 3.9ADD255 pKa = 3.41LGAAIDD261 pKa = 4.82AILDD265 pKa = 3.74EE266 pKa = 4.81EE267 pKa = 4.51IKK269 pKa = 10.59RR270 pKa = 11.84VEE272 pKa = 4.12GSQHH276 pKa = 6.93IEE278 pKa = 3.42FDD280 pKa = 3.77SYY282 pKa = 10.99QEE284 pKa = 3.82HH285 pKa = 6.96WNDD288 pKa = 2.68RR289 pKa = 11.84WAWAVNGAHH298 pKa = 6.48SGHH301 pKa = 6.12VSRR304 pKa = 11.84LYY306 pKa = 10.33PRR308 pKa = 11.84VPKK311 pKa = 10.14PPGMLRR317 pKa = 11.84EE318 pKa = 4.16HH319 pKa = 6.09RR320 pKa = 11.84RR321 pKa = 11.84AWLEE325 pKa = 3.93SVTEE329 pKa = 4.26DD330 pKa = 5.17PRR332 pKa = 11.84TDD334 pKa = 3.01WDD336 pKa = 3.47GKK338 pKa = 7.95TFVSASPKK346 pKa = 10.49LEE348 pKa = 4.2AGKK351 pKa = 8.26TRR353 pKa = 11.84AIFACDD359 pKa = 3.2TVNYY363 pKa = 10.17LAFEE367 pKa = 4.41HH368 pKa = 6.8LLAPVEE374 pKa = 4.2KK375 pKa = 9.55RR376 pKa = 11.84WRR378 pKa = 11.84NSKK381 pKa = 10.7VILDD385 pKa = 4.09PGRR388 pKa = 11.84GGHH391 pKa = 6.75LGMIFRR397 pKa = 11.84TTAARR402 pKa = 11.84ARR404 pKa = 11.84AGVSMMLDD412 pKa = 3.12YY413 pKa = 11.58DD414 pKa = 4.48DD415 pKa = 5.93FNSHH419 pKa = 6.2HH420 pKa = 6.27STRR423 pKa = 11.84AMQMLFQRR431 pKa = 11.84LGDD434 pKa = 3.7RR435 pKa = 11.84VGYY438 pKa = 8.83PADD441 pKa = 3.69KK442 pKa = 10.33LAKK445 pKa = 9.98LVASFEE451 pKa = 3.91KK452 pKa = 9.89MYY454 pKa = 10.0IYY456 pKa = 10.95NGMEE460 pKa = 3.43QVGRR464 pKa = 11.84VKK466 pKa = 9.33GTLMSGHH473 pKa = 7.16RR474 pKa = 11.84GTTFINSVLNKK485 pKa = 10.19AYY487 pKa = 10.86LLIVLGEE494 pKa = 4.19DD495 pKa = 3.0LFEE498 pKa = 5.48RR499 pKa = 11.84SVALHH504 pKa = 6.33VGDD507 pKa = 4.47DD508 pKa = 3.84VYY510 pKa = 11.4FGVRR514 pKa = 11.84TYY516 pKa = 11.59AEE518 pKa = 4.12AGEE521 pKa = 4.43VVTRR525 pKa = 11.84IKK527 pKa = 10.88NSPLRR532 pKa = 11.84MNRR535 pKa = 11.84MKK537 pKa = 10.71QSVGHH542 pKa = 6.0VSTEE546 pKa = 3.74FLRR549 pKa = 11.84NATSGRR555 pKa = 11.84STYY558 pKa = 10.86GYY560 pKa = 9.41FARR563 pKa = 11.84AVASTVSGNWVNEE576 pKa = 4.02MALSPSEE583 pKa = 4.05ALSSIIGAARR593 pKa = 11.84TLVNRR598 pKa = 11.84SGAVNLPLLLHH609 pKa = 6.56PSLVRR614 pKa = 11.84MTGLPRR620 pKa = 11.84EE621 pKa = 4.17DD622 pKa = 3.23HH623 pKa = 6.31KK624 pKa = 11.39KK625 pKa = 10.45LRR627 pKa = 11.84EE628 pKa = 4.04LLLGTTALDD637 pKa = 3.57NGPQYY642 pKa = 11.49SLGGYY647 pKa = 6.23YY648 pKa = 10.23TSVSSLITTTASDD661 pKa = 3.17RR662 pKa = 11.84HH663 pKa = 6.49GYY665 pKa = 7.63TPLPRR670 pKa = 11.84EE671 pKa = 3.83ATTAYY676 pKa = 9.93LSCAADD682 pKa = 3.59PLEE685 pKa = 4.54VNVLTQAGVSVVSAMEE701 pKa = 3.68EE702 pKa = 3.73ASFRR706 pKa = 11.84KK707 pKa = 9.57SLPARR712 pKa = 11.84YY713 pKa = 7.55TGYY716 pKa = 8.27EE717 pKa = 4.09TLRR720 pKa = 11.84LGPKK724 pKa = 9.94SLIRR728 pKa = 11.84SIGTASVVDD737 pKa = 5.93LISASPPRR745 pKa = 11.84GVLEE749 pKa = 4.66RR750 pKa = 11.84YY751 pKa = 8.65PLLTLAKK758 pKa = 9.7RR759 pKa = 11.84RR760 pKa = 11.84LPEE763 pKa = 3.81YY764 pKa = 9.59LVRR767 pKa = 11.84WAVSIAGGNPSAPDD781 pKa = 3.03IGMEE785 pKa = 3.47AWGEE789 pKa = 4.07FKK791 pKa = 10.36HH792 pKa = 6.83GCMIATPMSYY802 pKa = 10.89SDD804 pKa = 3.38AATFGKK810 pKa = 8.94RR811 pKa = 11.84TVSTVLTCPLDD822 pKa = 3.43AHH824 pKa = 6.15VV825 pKa = 4.57
MM1 pKa = 7.53IEE3 pKa = 4.23SPVSVRR9 pKa = 11.84ANEE12 pKa = 3.86AGIVGQYY19 pKa = 11.03LKK21 pKa = 11.21GLLNMEE27 pKa = 4.36WASGVMVLSFSQQISEE43 pKa = 4.43VYY45 pKa = 10.21KK46 pKa = 9.26PTFNGLRR53 pKa = 11.84PTDD56 pKa = 3.86LQRR59 pKa = 11.84AAAAYY64 pKa = 7.87LVPDD68 pKa = 4.48FPVQVRR74 pKa = 11.84IEE76 pKa = 4.17RR77 pKa = 11.84GSVLSLLSQVIDD89 pKa = 3.9PPARR93 pKa = 11.84VTDD96 pKa = 3.48RR97 pKa = 11.84CSFRR101 pKa = 11.84WLCDD105 pKa = 3.48PKK107 pKa = 11.01ASYY110 pKa = 10.66AAFPPKK116 pKa = 9.91QHH118 pKa = 6.89PGAMNKK124 pKa = 9.3VNVYY128 pKa = 9.74LNEE131 pKa = 4.12VASSLLSLDD140 pKa = 3.34PAAYY144 pKa = 10.2SSASQALWPHH154 pKa = 6.4KK155 pKa = 10.68GKK157 pKa = 10.17IANDD161 pKa = 3.31QASAIILYY169 pKa = 10.25GYY171 pKa = 8.74GLRR174 pKa = 11.84AQAVPDD180 pKa = 3.87AMHH183 pKa = 6.32VAATLATTPDD193 pKa = 3.35LAKK196 pKa = 10.91ALTNFLKK203 pKa = 10.29ATGANGSRR211 pKa = 11.84LGALLCEE218 pKa = 4.72SNVLLGRR225 pKa = 11.84AAGPADD231 pKa = 3.67LSEE234 pKa = 4.07EE235 pKa = 3.73ARR237 pKa = 11.84YY238 pKa = 7.94RR239 pKa = 11.84TSSDD243 pKa = 3.63VEE245 pKa = 3.92SHH247 pKa = 6.78LAIFSDD253 pKa = 3.9ADD255 pKa = 3.41LGAAIDD261 pKa = 4.82AILDD265 pKa = 3.74EE266 pKa = 4.81EE267 pKa = 4.51IKK269 pKa = 10.59RR270 pKa = 11.84VEE272 pKa = 4.12GSQHH276 pKa = 6.93IEE278 pKa = 3.42FDD280 pKa = 3.77SYY282 pKa = 10.99QEE284 pKa = 3.82HH285 pKa = 6.96WNDD288 pKa = 2.68RR289 pKa = 11.84WAWAVNGAHH298 pKa = 6.48SGHH301 pKa = 6.12VSRR304 pKa = 11.84LYY306 pKa = 10.33PRR308 pKa = 11.84VPKK311 pKa = 10.14PPGMLRR317 pKa = 11.84EE318 pKa = 4.16HH319 pKa = 6.09RR320 pKa = 11.84RR321 pKa = 11.84AWLEE325 pKa = 3.93SVTEE329 pKa = 4.26DD330 pKa = 5.17PRR332 pKa = 11.84TDD334 pKa = 3.01WDD336 pKa = 3.47GKK338 pKa = 7.95TFVSASPKK346 pKa = 10.49LEE348 pKa = 4.2AGKK351 pKa = 8.26TRR353 pKa = 11.84AIFACDD359 pKa = 3.2TVNYY363 pKa = 10.17LAFEE367 pKa = 4.41HH368 pKa = 6.8LLAPVEE374 pKa = 4.2KK375 pKa = 9.55RR376 pKa = 11.84WRR378 pKa = 11.84NSKK381 pKa = 10.7VILDD385 pKa = 4.09PGRR388 pKa = 11.84GGHH391 pKa = 6.75LGMIFRR397 pKa = 11.84TTAARR402 pKa = 11.84ARR404 pKa = 11.84AGVSMMLDD412 pKa = 3.12YY413 pKa = 11.58DD414 pKa = 4.48DD415 pKa = 5.93FNSHH419 pKa = 6.2HH420 pKa = 6.27STRR423 pKa = 11.84AMQMLFQRR431 pKa = 11.84LGDD434 pKa = 3.7RR435 pKa = 11.84VGYY438 pKa = 8.83PADD441 pKa = 3.69KK442 pKa = 10.33LAKK445 pKa = 9.98LVASFEE451 pKa = 3.91KK452 pKa = 9.89MYY454 pKa = 10.0IYY456 pKa = 10.95NGMEE460 pKa = 3.43QVGRR464 pKa = 11.84VKK466 pKa = 9.33GTLMSGHH473 pKa = 7.16RR474 pKa = 11.84GTTFINSVLNKK485 pKa = 10.19AYY487 pKa = 10.86LLIVLGEE494 pKa = 4.19DD495 pKa = 3.0LFEE498 pKa = 5.48RR499 pKa = 11.84SVALHH504 pKa = 6.33VGDD507 pKa = 4.47DD508 pKa = 3.84VYY510 pKa = 11.4FGVRR514 pKa = 11.84TYY516 pKa = 11.59AEE518 pKa = 4.12AGEE521 pKa = 4.43VVTRR525 pKa = 11.84IKK527 pKa = 10.88NSPLRR532 pKa = 11.84MNRR535 pKa = 11.84MKK537 pKa = 10.71QSVGHH542 pKa = 6.0VSTEE546 pKa = 3.74FLRR549 pKa = 11.84NATSGRR555 pKa = 11.84STYY558 pKa = 10.86GYY560 pKa = 9.41FARR563 pKa = 11.84AVASTVSGNWVNEE576 pKa = 4.02MALSPSEE583 pKa = 4.05ALSSIIGAARR593 pKa = 11.84TLVNRR598 pKa = 11.84SGAVNLPLLLHH609 pKa = 6.56PSLVRR614 pKa = 11.84MTGLPRR620 pKa = 11.84EE621 pKa = 4.17DD622 pKa = 3.23HH623 pKa = 6.31KK624 pKa = 11.39KK625 pKa = 10.45LRR627 pKa = 11.84EE628 pKa = 4.04LLLGTTALDD637 pKa = 3.57NGPQYY642 pKa = 11.49SLGGYY647 pKa = 6.23YY648 pKa = 10.23TSVSSLITTTASDD661 pKa = 3.17RR662 pKa = 11.84HH663 pKa = 6.49GYY665 pKa = 7.63TPLPRR670 pKa = 11.84EE671 pKa = 3.83ATTAYY676 pKa = 9.93LSCAADD682 pKa = 3.59PLEE685 pKa = 4.54VNVLTQAGVSVVSAMEE701 pKa = 3.68EE702 pKa = 3.73ASFRR706 pKa = 11.84KK707 pKa = 9.57SLPARR712 pKa = 11.84YY713 pKa = 7.55TGYY716 pKa = 8.27EE717 pKa = 4.09TLRR720 pKa = 11.84LGPKK724 pKa = 9.94SLIRR728 pKa = 11.84SIGTASVVDD737 pKa = 5.93LISASPPRR745 pKa = 11.84GVLEE749 pKa = 4.66RR750 pKa = 11.84YY751 pKa = 8.65PLLTLAKK758 pKa = 9.7RR759 pKa = 11.84RR760 pKa = 11.84LPEE763 pKa = 3.81YY764 pKa = 9.59LVRR767 pKa = 11.84WAVSIAGGNPSAPDD781 pKa = 3.03IGMEE785 pKa = 3.47AWGEE789 pKa = 4.07FKK791 pKa = 10.36HH792 pKa = 6.83GCMIATPMSYY802 pKa = 10.89SDD804 pKa = 3.38AATFGKK810 pKa = 8.94RR811 pKa = 11.84TVSTVLTCPLDD822 pKa = 3.43AHH824 pKa = 6.15VV825 pKa = 4.57
Molecular weight: 90.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1601 |
776 |
825 |
800.5 |
85.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.554 ± 1.778 | 0.812 ± 0.027 |
4.872 ± 0.197 | 4.435 ± 0.396 |
3.123 ± 0.248 | 8.307 ± 0.676 |
2.561 ± 0.011 | 3.248 ± 0.198 |
2.561 ± 0.796 | 8.62 ± 1.426 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.249 ± 0.309 | 3.435 ± 0.031 |
6.996 ± 1.23 | 2.498 ± 0.234 |
6.558 ± 0.349 | 7.808 ± 0.77 |
6.246 ± 0.406 | 7.995 ± 0.265 |
1.187 ± 0.198 | 2.936 ± 0.608 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
