
Chicken stool-associated gemycircularvirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykrogvirus; Gemykrogvirus galga2
Average proteome isoelectric point is 8.77
Get precalculated fractions of proteins
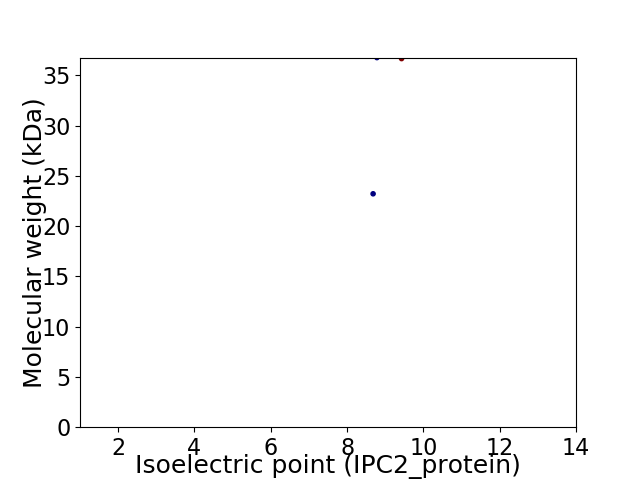
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L6KW52|A0A1L6KW52_9VIRU Capsid protein OS=Chicken stool-associated gemycircularvirus OX=1930298 PE=4 SV=1
MM1 pKa = 6.97TRR3 pKa = 11.84FRR5 pKa = 11.84LRR7 pKa = 11.84AKK9 pKa = 10.2IFFLTYY15 pKa = 9.88SQIGEE20 pKa = 4.22DD21 pKa = 3.93VIQEE25 pKa = 4.06LRR27 pKa = 11.84STPTQHH33 pKa = 6.22FRR35 pKa = 11.84FIEE38 pKa = 4.3EE39 pKa = 4.18VLGTPSDD46 pKa = 3.75YY47 pKa = 10.65RR48 pKa = 11.84LSRR51 pKa = 11.84EE52 pKa = 3.93SHH54 pKa = 5.87QDD56 pKa = 3.22GASHH60 pKa = 6.11LHH62 pKa = 6.05CVVGYY67 pKa = 11.22AEE69 pKa = 4.23TQLVNSASLFDD80 pKa = 3.86YY81 pKa = 10.78RR82 pKa = 11.84GAHH85 pKa = 6.31PNIKK89 pKa = 10.01SIRR92 pKa = 11.84GTIKK96 pKa = 10.75KK97 pKa = 8.29PWNYY101 pKa = 10.43AGKK104 pKa = 10.17DD105 pKa = 3.04GDD107 pKa = 4.55IIHH110 pKa = 6.86EE111 pKa = 4.36LGGPNGGDD119 pKa = 3.11SGTGGSIAEE128 pKa = 4.23RR129 pKa = 11.84WSHH132 pKa = 7.12AIDD135 pKa = 4.55APTEE139 pKa = 3.97DD140 pKa = 3.72EE141 pKa = 4.03FFEE144 pKa = 4.44RR145 pKa = 11.84VRR147 pKa = 11.84MADD150 pKa = 3.08PRR152 pKa = 11.84SYY154 pKa = 11.68VLFNRR159 pKa = 11.84AIVQYY164 pKa = 11.01ANKK167 pKa = 9.77IYY169 pKa = 10.33RR170 pKa = 11.84KK171 pKa = 8.14PRR173 pKa = 11.84EE174 pKa = 3.89YY175 pKa = 10.01TSPRR179 pKa = 11.84FRR181 pKa = 11.84LLGGSRR187 pKa = 11.84IEE189 pKa = 4.24GWLSQSDD196 pKa = 3.43LGRR199 pKa = 11.84RR200 pKa = 11.84GGEE203 pKa = 3.74RR204 pKa = 3.19
MM1 pKa = 6.97TRR3 pKa = 11.84FRR5 pKa = 11.84LRR7 pKa = 11.84AKK9 pKa = 10.2IFFLTYY15 pKa = 9.88SQIGEE20 pKa = 4.22DD21 pKa = 3.93VIQEE25 pKa = 4.06LRR27 pKa = 11.84STPTQHH33 pKa = 6.22FRR35 pKa = 11.84FIEE38 pKa = 4.3EE39 pKa = 4.18VLGTPSDD46 pKa = 3.75YY47 pKa = 10.65RR48 pKa = 11.84LSRR51 pKa = 11.84EE52 pKa = 3.93SHH54 pKa = 5.87QDD56 pKa = 3.22GASHH60 pKa = 6.11LHH62 pKa = 6.05CVVGYY67 pKa = 11.22AEE69 pKa = 4.23TQLVNSASLFDD80 pKa = 3.86YY81 pKa = 10.78RR82 pKa = 11.84GAHH85 pKa = 6.31PNIKK89 pKa = 10.01SIRR92 pKa = 11.84GTIKK96 pKa = 10.75KK97 pKa = 8.29PWNYY101 pKa = 10.43AGKK104 pKa = 10.17DD105 pKa = 3.04GDD107 pKa = 4.55IIHH110 pKa = 6.86EE111 pKa = 4.36LGGPNGGDD119 pKa = 3.11SGTGGSIAEE128 pKa = 4.23RR129 pKa = 11.84WSHH132 pKa = 7.12AIDD135 pKa = 4.55APTEE139 pKa = 3.97DD140 pKa = 3.72EE141 pKa = 4.03FFEE144 pKa = 4.44RR145 pKa = 11.84VRR147 pKa = 11.84MADD150 pKa = 3.08PRR152 pKa = 11.84SYY154 pKa = 11.68VLFNRR159 pKa = 11.84AIVQYY164 pKa = 11.01ANKK167 pKa = 9.77IYY169 pKa = 10.33RR170 pKa = 11.84KK171 pKa = 8.14PRR173 pKa = 11.84EE174 pKa = 3.89YY175 pKa = 10.01TSPRR179 pKa = 11.84FRR181 pKa = 11.84LLGGSRR187 pKa = 11.84IEE189 pKa = 4.24GWLSQSDD196 pKa = 3.43LGRR199 pKa = 11.84RR200 pKa = 11.84GGEE203 pKa = 3.74RR204 pKa = 3.19
Molecular weight: 23.21 kDa
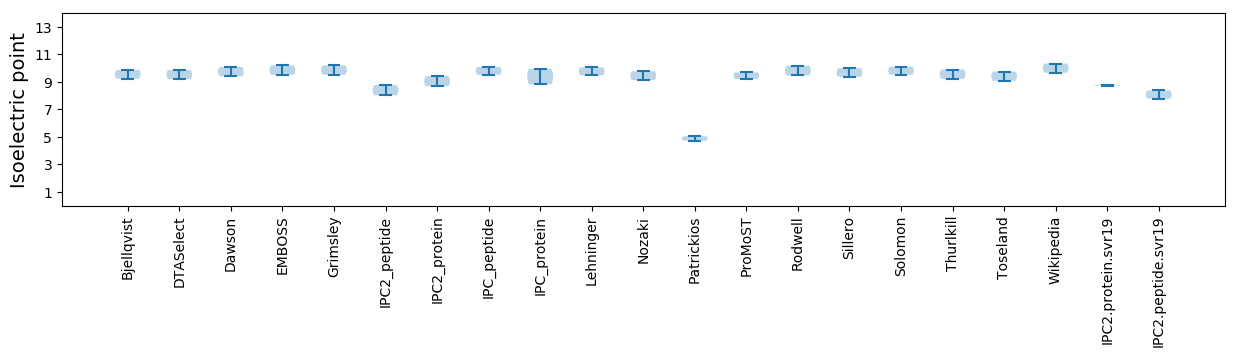
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L6KW52|A0A1L6KW52_9VIRU Capsid protein OS=Chicken stool-associated gemycircularvirus OX=1930298 PE=4 SV=1
MM1 pKa = 7.84GYY3 pKa = 10.16FEE5 pKa = 5.2EE6 pKa = 4.69NGYY9 pKa = 10.11SLSTGLHH16 pKa = 5.85HH17 pKa = 7.91SEE19 pKa = 3.51MAYY22 pKa = 10.37SRR24 pKa = 11.84FRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84SFRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84PARR37 pKa = 11.84TLRR40 pKa = 11.84RR41 pKa = 11.84SQHH44 pKa = 4.73KK45 pKa = 8.15RR46 pKa = 11.84TVRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84FVRR55 pKa = 11.84RR56 pKa = 11.84PRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84MTSRR64 pKa = 11.84RR65 pKa = 11.84VRR67 pKa = 11.84NIASRR72 pKa = 11.84KK73 pKa = 9.04KK74 pKa = 10.06HH75 pKa = 5.38DD76 pKa = 3.74TQLGADD82 pKa = 3.93SNADD86 pKa = 3.42DD87 pKa = 4.64SSNPVQLHH95 pKa = 6.38AGNNYY100 pKa = 8.68YY101 pKa = 10.68LWAPSYY107 pKa = 10.85LEE109 pKa = 3.86QHH111 pKa = 6.2EE112 pKa = 4.22EE113 pKa = 3.96SYY115 pKa = 11.36DD116 pKa = 3.7YY117 pKa = 11.45VRR119 pKa = 11.84NNQNVYY125 pKa = 10.06FRR127 pKa = 11.84GVQDD131 pKa = 3.94RR132 pKa = 11.84VFLSAAFAFTWRR144 pKa = 11.84RR145 pKa = 11.84VCFFSYY151 pKa = 10.5EE152 pKa = 3.98QFIPALPFFVEE163 pKa = 5.26APNEE167 pKa = 4.16DD168 pKa = 4.02DD169 pKa = 3.73PQIMRR174 pKa = 11.84RR175 pKa = 11.84PLRR178 pKa = 11.84EE179 pKa = 3.91ITPKK183 pKa = 9.38TASEE187 pKa = 4.2FFSYY191 pKa = 9.82AWKK194 pKa = 8.9GTVGVDD200 pKa = 3.83FSEE203 pKa = 4.13NTRR206 pKa = 11.84WNAPLDD212 pKa = 4.29DD213 pKa = 4.5KK214 pKa = 11.5LLTIVYY220 pKa = 9.6DD221 pKa = 3.84RR222 pKa = 11.84SVTINPNYY230 pKa = 9.75AAPAGATFGKK240 pKa = 10.8SMTRR244 pKa = 11.84KK245 pKa = 8.12MWHH248 pKa = 6.45PVNKK252 pKa = 9.83TMMYY256 pKa = 10.12SDD258 pKa = 4.9KK259 pKa = 11.03EE260 pKa = 4.23NGSTPSSTGWTSMCPNSPGNFYY282 pKa = 10.64ILDD285 pKa = 3.84IFSTGQDD292 pKa = 3.42LPGDD296 pKa = 3.67LTGVGEE302 pKa = 4.68FGSEE306 pKa = 3.61ATVYY310 pKa = 8.37WHH312 pKa = 6.95EE313 pKa = 4.1NN314 pKa = 3.13
MM1 pKa = 7.84GYY3 pKa = 10.16FEE5 pKa = 5.2EE6 pKa = 4.69NGYY9 pKa = 10.11SLSTGLHH16 pKa = 5.85HH17 pKa = 7.91SEE19 pKa = 3.51MAYY22 pKa = 10.37SRR24 pKa = 11.84FRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84SFRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84PARR37 pKa = 11.84TLRR40 pKa = 11.84RR41 pKa = 11.84SQHH44 pKa = 4.73KK45 pKa = 8.15RR46 pKa = 11.84TVRR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84FVRR55 pKa = 11.84RR56 pKa = 11.84PRR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84MTSRR64 pKa = 11.84RR65 pKa = 11.84VRR67 pKa = 11.84NIASRR72 pKa = 11.84KK73 pKa = 9.04KK74 pKa = 10.06HH75 pKa = 5.38DD76 pKa = 3.74TQLGADD82 pKa = 3.93SNADD86 pKa = 3.42DD87 pKa = 4.64SSNPVQLHH95 pKa = 6.38AGNNYY100 pKa = 8.68YY101 pKa = 10.68LWAPSYY107 pKa = 10.85LEE109 pKa = 3.86QHH111 pKa = 6.2EE112 pKa = 4.22EE113 pKa = 3.96SYY115 pKa = 11.36DD116 pKa = 3.7YY117 pKa = 11.45VRR119 pKa = 11.84NNQNVYY125 pKa = 10.06FRR127 pKa = 11.84GVQDD131 pKa = 3.94RR132 pKa = 11.84VFLSAAFAFTWRR144 pKa = 11.84RR145 pKa = 11.84VCFFSYY151 pKa = 10.5EE152 pKa = 3.98QFIPALPFFVEE163 pKa = 5.26APNEE167 pKa = 4.16DD168 pKa = 4.02DD169 pKa = 3.73PQIMRR174 pKa = 11.84RR175 pKa = 11.84PLRR178 pKa = 11.84EE179 pKa = 3.91ITPKK183 pKa = 9.38TASEE187 pKa = 4.2FFSYY191 pKa = 9.82AWKK194 pKa = 8.9GTVGVDD200 pKa = 3.83FSEE203 pKa = 4.13NTRR206 pKa = 11.84WNAPLDD212 pKa = 4.29DD213 pKa = 4.5KK214 pKa = 11.5LLTIVYY220 pKa = 9.6DD221 pKa = 3.84RR222 pKa = 11.84SVTINPNYY230 pKa = 9.75AAPAGATFGKK240 pKa = 10.8SMTRR244 pKa = 11.84KK245 pKa = 8.12MWHH248 pKa = 6.45PVNKK252 pKa = 9.83TMMYY256 pKa = 10.12SDD258 pKa = 4.9KK259 pKa = 11.03EE260 pKa = 4.23NGSTPSSTGWTSMCPNSPGNFYY282 pKa = 10.64ILDD285 pKa = 3.84IFSTGQDD292 pKa = 3.42LPGDD296 pKa = 3.67LTGVGEE302 pKa = 4.68FGSEE306 pKa = 3.61ATVYY310 pKa = 8.37WHH312 pKa = 6.95EE313 pKa = 4.1NN314 pKa = 3.13
Molecular weight: 36.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
518 |
204 |
314 |
259.0 |
29.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.178 ± 0.176 | 0.579 ± 0.053 |
5.212 ± 0.107 | 5.792 ± 0.637 |
5.792 ± 0.529 | 7.529 ± 1.645 |
2.896 ± 0.319 | 4.247 ± 1.556 |
3.282 ± 0.089 | 5.792 ± 0.637 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.124 ± 0.68 | 4.826 ± 1.122 |
5.212 ± 0.476 | 3.089 ± 0.204 |
11.197 ± 0.246 | 8.687 ± 0.211 |
5.985 ± 0.644 | 4.826 ± 0.538 |
1.931 ± 0.274 | 4.826 ± 0.247 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
