
Wenzhou tombus-like virus 10
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.68
Get precalculated fractions of proteins
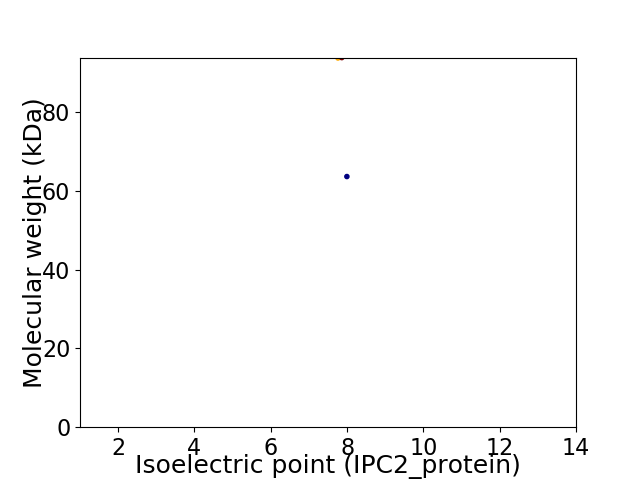
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KH44|A0A1L3KH44_9VIRU Uncharacterized protein OS=Wenzhou tombus-like virus 10 OX=1923663 PE=4 SV=1
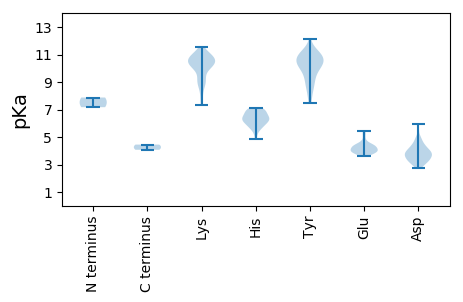
MM1 pKa = 7.87TDD3 pKa = 3.12QQFHH7 pKa = 6.39MLPKK11 pKa = 10.41LVRR14 pKa = 11.84QFVLRR19 pKa = 11.84RR20 pKa = 11.84VRR22 pKa = 11.84TTIRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84CVNRR32 pKa = 11.84GRR34 pKa = 11.84GKK36 pKa = 10.81LLGNKK41 pKa = 8.15MMHH44 pKa = 6.83ALNGNTTDD52 pKa = 3.67YY53 pKa = 11.36VKK55 pKa = 10.22WKK57 pKa = 10.46QSFDD61 pKa = 3.52EE62 pKa = 4.4SLKK65 pKa = 7.86TTNDD69 pKa = 2.76KK70 pKa = 10.95SRR72 pKa = 11.84QQRR75 pKa = 11.84VDD77 pKa = 3.0MNIRR81 pKa = 11.84VSVDD85 pKa = 3.14MPKK88 pKa = 9.48TFSLTHH94 pKa = 6.92DD95 pKa = 4.4LAAMAFVAVLDD106 pKa = 3.62GFMYY110 pKa = 10.57HH111 pKa = 7.01IAFDD115 pKa = 3.89YY116 pKa = 10.73LKK118 pKa = 10.36KK119 pKa = 10.84GKK121 pKa = 9.79AKK123 pKa = 10.37RR124 pKa = 11.84IIVEE128 pKa = 4.05MPHH131 pKa = 6.51PVNGCTKK138 pKa = 10.02QCGWDD143 pKa = 3.69DD144 pKa = 3.77NVVRR148 pKa = 11.84SYY150 pKa = 10.26PAPLLINDD158 pKa = 5.51IIPIPLPCEE167 pKa = 3.84VTFGRR172 pKa = 11.84NGLQKK177 pKa = 10.73HH178 pKa = 6.57FITDD182 pKa = 3.09DD183 pKa = 3.81DD184 pKa = 4.39YY185 pKa = 12.13ADD187 pKa = 4.09FDD189 pKa = 4.35IPLDD193 pKa = 4.13DD194 pKa = 4.53SLSNLQYY201 pKa = 11.44DD202 pKa = 4.11SDD204 pKa = 4.09TKK206 pKa = 10.67SWNSAGSPAQSTSSNVSGVTAPSSSTPTPSTSAPPPKK243 pKa = 10.18SNKK246 pKa = 8.52QLSIVRR252 pKa = 11.84RR253 pKa = 11.84SDD255 pKa = 3.43SKK257 pKa = 10.55VQTPSVEE264 pKa = 3.75QRR266 pKa = 11.84VCVVPPLQPTPSQPTPPPTLAAPRR290 pKa = 11.84VVSEE294 pKa = 5.35GYY296 pKa = 8.56TEE298 pKa = 3.92PHH300 pKa = 5.67TRR302 pKa = 11.84LHH304 pKa = 5.95FLARR308 pKa = 11.84HH309 pKa = 5.97AAQISEE315 pKa = 4.44VEE317 pKa = 4.45RR318 pKa = 11.84PQLPDD323 pKa = 3.26FTEE326 pKa = 3.77NPYY329 pKa = 10.44RR330 pKa = 11.84KK331 pKa = 8.73WVAEE335 pKa = 3.92VLLGDD340 pKa = 3.86RR341 pKa = 11.84NNPNMQFPLLDD352 pKa = 3.75LRR354 pKa = 11.84SPLAIWMSSGSFKK367 pKa = 10.71TVMRR371 pKa = 11.84KK372 pKa = 9.85HH373 pKa = 5.27IQYY376 pKa = 9.53GNSMGLDD383 pKa = 3.27AEE385 pKa = 4.47MLVHH389 pKa = 6.89LVRR392 pKa = 11.84LITEE396 pKa = 3.86KK397 pKa = 10.95RR398 pKa = 11.84EE399 pKa = 3.97ASSFATLYY407 pKa = 10.47HH408 pKa = 7.08KK409 pKa = 10.72AYY411 pKa = 8.76TWASHH416 pKa = 6.21NRR418 pKa = 11.84EE419 pKa = 4.22DD420 pKa = 3.98WPLGLSDD427 pKa = 4.34AQSAGCARR435 pKa = 11.84YY436 pKa = 7.7TLGTSSLYY444 pKa = 10.9ASLLNNITPGVSEE457 pKa = 4.49EE458 pKa = 4.42SFDD461 pKa = 4.45VDD463 pKa = 3.53PQINPVYY470 pKa = 9.25ATKK473 pKa = 10.5NRR475 pKa = 11.84RR476 pKa = 11.84NFVYY480 pKa = 10.47NVVLIILFILFSYY493 pKa = 9.97AAYY496 pKa = 8.68HH497 pKa = 5.66TFRR500 pKa = 11.84FARR503 pKa = 11.84YY504 pKa = 9.53VDD506 pKa = 2.94GHH508 pKa = 5.56YY509 pKa = 11.2NRR511 pKa = 11.84FAAYY515 pKa = 10.24ADD517 pKa = 3.42NSFEE521 pKa = 4.25NFGNQFTAFAAVTYY535 pKa = 10.21GSSCFTHH542 pKa = 6.42EE543 pKa = 5.45RR544 pKa = 11.84IGCALGDD551 pKa = 3.57NDD553 pKa = 4.96CYY555 pKa = 10.97RR556 pKa = 11.84RR557 pKa = 11.84NCRR560 pKa = 11.84DD561 pKa = 3.22LLRR564 pKa = 11.84IYY566 pKa = 10.75HH567 pKa = 6.92PDD569 pKa = 2.84KK570 pKa = 11.09GGNVDD575 pKa = 3.73SYY577 pKa = 9.07THH579 pKa = 5.7VARR582 pKa = 11.84DD583 pKa = 3.82CEE585 pKa = 4.19FLNKK589 pKa = 10.31GGEE592 pKa = 4.03QPVYY596 pKa = 10.42EE597 pKa = 4.22NCAYY601 pKa = 10.4ANKK604 pKa = 10.23EE605 pKa = 3.92EE606 pKa = 4.4GVSFEE611 pKa = 4.8LNEE614 pKa = 4.11LYY616 pKa = 11.06VRR618 pKa = 11.84FSQFAKK624 pKa = 10.12TRR626 pKa = 11.84GIDD629 pKa = 3.56ADD631 pKa = 4.07FGDD634 pKa = 3.94YY635 pKa = 10.8DD636 pKa = 4.48YY637 pKa = 11.2FFQAAPSSNPICDD650 pKa = 3.05AVKK653 pKa = 10.66NGEE656 pKa = 4.34SVPKK660 pKa = 8.98EE661 pKa = 3.77VLMICQFFKK670 pKa = 11.13LRR672 pKa = 11.84DD673 pKa = 3.96FRR675 pKa = 11.84PSCYY679 pKa = 10.04NYY681 pKa = 10.69SSFFNAYY688 pKa = 9.56GSMYY692 pKa = 10.06HH693 pKa = 6.31NGIIFQSIFPTMVSICVSFGKK714 pKa = 10.02GQTMRR719 pKa = 11.84EE720 pKa = 3.88HH721 pKa = 6.67MFTEE725 pKa = 5.19VYY727 pKa = 9.17YY728 pKa = 10.83TMAFQYY734 pKa = 10.88CCVCCFVALISVLFATLAIYY754 pKa = 7.49PTVKK758 pKa = 10.16KK759 pKa = 10.85AFIEE763 pKa = 3.87RR764 pKa = 11.84GFRR767 pKa = 11.84VGCSVWVSKK776 pKa = 11.02CSVTGVTSAAIMATFILIKK795 pKa = 10.47EE796 pKa = 4.04RR797 pKa = 11.84DD798 pKa = 3.93LLCRR802 pKa = 11.84ASSAAYY808 pKa = 9.23VARR811 pKa = 11.84ALKK814 pKa = 10.27EE815 pKa = 3.78LLLLVISGLRR825 pKa = 11.84SLWAAAAAA833 pKa = 4.07
MM1 pKa = 7.87TDD3 pKa = 3.12QQFHH7 pKa = 6.39MLPKK11 pKa = 10.41LVRR14 pKa = 11.84QFVLRR19 pKa = 11.84RR20 pKa = 11.84VRR22 pKa = 11.84TTIRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84CVNRR32 pKa = 11.84GRR34 pKa = 11.84GKK36 pKa = 10.81LLGNKK41 pKa = 8.15MMHH44 pKa = 6.83ALNGNTTDD52 pKa = 3.67YY53 pKa = 11.36VKK55 pKa = 10.22WKK57 pKa = 10.46QSFDD61 pKa = 3.52EE62 pKa = 4.4SLKK65 pKa = 7.86TTNDD69 pKa = 2.76KK70 pKa = 10.95SRR72 pKa = 11.84QQRR75 pKa = 11.84VDD77 pKa = 3.0MNIRR81 pKa = 11.84VSVDD85 pKa = 3.14MPKK88 pKa = 9.48TFSLTHH94 pKa = 6.92DD95 pKa = 4.4LAAMAFVAVLDD106 pKa = 3.62GFMYY110 pKa = 10.57HH111 pKa = 7.01IAFDD115 pKa = 3.89YY116 pKa = 10.73LKK118 pKa = 10.36KK119 pKa = 10.84GKK121 pKa = 9.79AKK123 pKa = 10.37RR124 pKa = 11.84IIVEE128 pKa = 4.05MPHH131 pKa = 6.51PVNGCTKK138 pKa = 10.02QCGWDD143 pKa = 3.69DD144 pKa = 3.77NVVRR148 pKa = 11.84SYY150 pKa = 10.26PAPLLINDD158 pKa = 5.51IIPIPLPCEE167 pKa = 3.84VTFGRR172 pKa = 11.84NGLQKK177 pKa = 10.73HH178 pKa = 6.57FITDD182 pKa = 3.09DD183 pKa = 3.81DD184 pKa = 4.39YY185 pKa = 12.13ADD187 pKa = 4.09FDD189 pKa = 4.35IPLDD193 pKa = 4.13DD194 pKa = 4.53SLSNLQYY201 pKa = 11.44DD202 pKa = 4.11SDD204 pKa = 4.09TKK206 pKa = 10.67SWNSAGSPAQSTSSNVSGVTAPSSSTPTPSTSAPPPKK243 pKa = 10.18SNKK246 pKa = 8.52QLSIVRR252 pKa = 11.84RR253 pKa = 11.84SDD255 pKa = 3.43SKK257 pKa = 10.55VQTPSVEE264 pKa = 3.75QRR266 pKa = 11.84VCVVPPLQPTPSQPTPPPTLAAPRR290 pKa = 11.84VVSEE294 pKa = 5.35GYY296 pKa = 8.56TEE298 pKa = 3.92PHH300 pKa = 5.67TRR302 pKa = 11.84LHH304 pKa = 5.95FLARR308 pKa = 11.84HH309 pKa = 5.97AAQISEE315 pKa = 4.44VEE317 pKa = 4.45RR318 pKa = 11.84PQLPDD323 pKa = 3.26FTEE326 pKa = 3.77NPYY329 pKa = 10.44RR330 pKa = 11.84KK331 pKa = 8.73WVAEE335 pKa = 3.92VLLGDD340 pKa = 3.86RR341 pKa = 11.84NNPNMQFPLLDD352 pKa = 3.75LRR354 pKa = 11.84SPLAIWMSSGSFKK367 pKa = 10.71TVMRR371 pKa = 11.84KK372 pKa = 9.85HH373 pKa = 5.27IQYY376 pKa = 9.53GNSMGLDD383 pKa = 3.27AEE385 pKa = 4.47MLVHH389 pKa = 6.89LVRR392 pKa = 11.84LITEE396 pKa = 3.86KK397 pKa = 10.95RR398 pKa = 11.84EE399 pKa = 3.97ASSFATLYY407 pKa = 10.47HH408 pKa = 7.08KK409 pKa = 10.72AYY411 pKa = 8.76TWASHH416 pKa = 6.21NRR418 pKa = 11.84EE419 pKa = 4.22DD420 pKa = 3.98WPLGLSDD427 pKa = 4.34AQSAGCARR435 pKa = 11.84YY436 pKa = 7.7TLGTSSLYY444 pKa = 10.9ASLLNNITPGVSEE457 pKa = 4.49EE458 pKa = 4.42SFDD461 pKa = 4.45VDD463 pKa = 3.53PQINPVYY470 pKa = 9.25ATKK473 pKa = 10.5NRR475 pKa = 11.84RR476 pKa = 11.84NFVYY480 pKa = 10.47NVVLIILFILFSYY493 pKa = 9.97AAYY496 pKa = 8.68HH497 pKa = 5.66TFRR500 pKa = 11.84FARR503 pKa = 11.84YY504 pKa = 9.53VDD506 pKa = 2.94GHH508 pKa = 5.56YY509 pKa = 11.2NRR511 pKa = 11.84FAAYY515 pKa = 10.24ADD517 pKa = 3.42NSFEE521 pKa = 4.25NFGNQFTAFAAVTYY535 pKa = 10.21GSSCFTHH542 pKa = 6.42EE543 pKa = 5.45RR544 pKa = 11.84IGCALGDD551 pKa = 3.57NDD553 pKa = 4.96CYY555 pKa = 10.97RR556 pKa = 11.84RR557 pKa = 11.84NCRR560 pKa = 11.84DD561 pKa = 3.22LLRR564 pKa = 11.84IYY566 pKa = 10.75HH567 pKa = 6.92PDD569 pKa = 2.84KK570 pKa = 11.09GGNVDD575 pKa = 3.73SYY577 pKa = 9.07THH579 pKa = 5.7VARR582 pKa = 11.84DD583 pKa = 3.82CEE585 pKa = 4.19FLNKK589 pKa = 10.31GGEE592 pKa = 4.03QPVYY596 pKa = 10.42EE597 pKa = 4.22NCAYY601 pKa = 10.4ANKK604 pKa = 10.23EE605 pKa = 3.92EE606 pKa = 4.4GVSFEE611 pKa = 4.8LNEE614 pKa = 4.11LYY616 pKa = 11.06VRR618 pKa = 11.84FSQFAKK624 pKa = 10.12TRR626 pKa = 11.84GIDD629 pKa = 3.56ADD631 pKa = 4.07FGDD634 pKa = 3.94YY635 pKa = 10.8DD636 pKa = 4.48YY637 pKa = 11.2FFQAAPSSNPICDD650 pKa = 3.05AVKK653 pKa = 10.66NGEE656 pKa = 4.34SVPKK660 pKa = 8.98EE661 pKa = 3.77VLMICQFFKK670 pKa = 11.13LRR672 pKa = 11.84DD673 pKa = 3.96FRR675 pKa = 11.84PSCYY679 pKa = 10.04NYY681 pKa = 10.69SSFFNAYY688 pKa = 9.56GSMYY692 pKa = 10.06HH693 pKa = 6.31NGIIFQSIFPTMVSICVSFGKK714 pKa = 10.02GQTMRR719 pKa = 11.84EE720 pKa = 3.88HH721 pKa = 6.67MFTEE725 pKa = 5.19VYY727 pKa = 9.17YY728 pKa = 10.83TMAFQYY734 pKa = 10.88CCVCCFVALISVLFATLAIYY754 pKa = 7.49PTVKK758 pKa = 10.16KK759 pKa = 10.85AFIEE763 pKa = 3.87RR764 pKa = 11.84GFRR767 pKa = 11.84VGCSVWVSKK776 pKa = 11.02CSVTGVTSAAIMATFILIKK795 pKa = 10.47EE796 pKa = 4.04RR797 pKa = 11.84DD798 pKa = 3.93LLCRR802 pKa = 11.84ASSAAYY808 pKa = 9.23VARR811 pKa = 11.84ALKK814 pKa = 10.27EE815 pKa = 3.78LLLLVISGLRR825 pKa = 11.84SLWAAAAAA833 pKa = 4.07
Molecular weight: 93.82 kDa
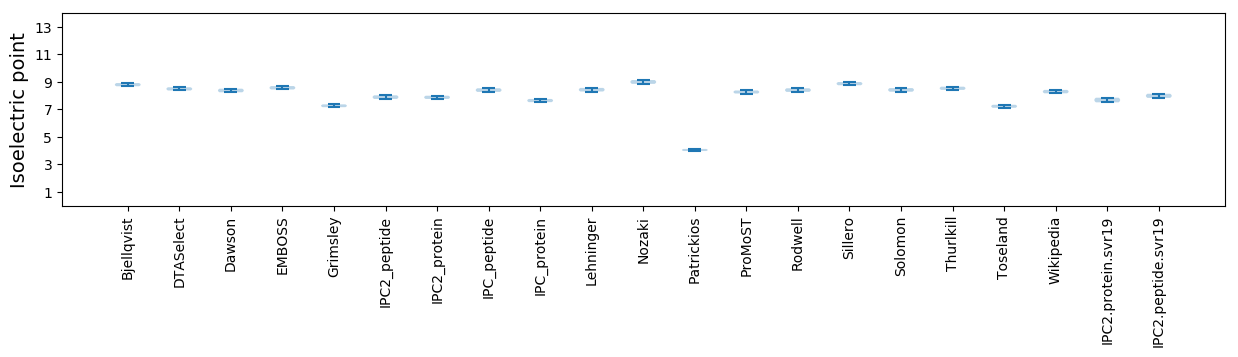
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KH44|A0A1L3KH44_9VIRU Uncharacterized protein OS=Wenzhou tombus-like virus 10 OX=1923663 PE=4 SV=1
MM1 pKa = 7.17FGYY4 pKa = 9.99RR5 pKa = 11.84RR6 pKa = 11.84HH7 pKa = 6.09ICSHH11 pKa = 6.6HH12 pKa = 6.69GNLHH16 pKa = 5.8TYY18 pKa = 7.89QRR20 pKa = 11.84EE21 pKa = 3.91RR22 pKa = 11.84SIVPCVVSSVCSSSSEE38 pKa = 3.9GAVIVGNKK46 pKa = 7.3WVAQPLGSGCSMSRR60 pKa = 11.84TNQQSEE66 pKa = 4.36SCSAKK71 pKa = 9.11FCKK74 pKa = 10.15FLTIKK79 pKa = 10.59NSRR82 pKa = 11.84TWDD85 pKa = 4.28PIVFSPCSHH94 pKa = 6.33NFMLAAKK101 pKa = 10.09SRR103 pKa = 11.84ILKK106 pKa = 7.52EE107 pKa = 4.22TPQPEE112 pKa = 4.09DD113 pKa = 3.29EE114 pKa = 4.31YY115 pKa = 11.47LRR117 pKa = 11.84RR118 pKa = 11.84FALSLHH124 pKa = 6.17AVAKK128 pKa = 10.47SIFVKK133 pKa = 8.71QTPLTRR139 pKa = 11.84EE140 pKa = 4.1EE141 pKa = 4.11FLSAYY146 pKa = 10.15SGSKK150 pKa = 9.11RR151 pKa = 11.84LRR153 pKa = 11.84YY154 pKa = 10.25AMAADD159 pKa = 3.66RR160 pKa = 11.84VYY162 pKa = 11.21DD163 pKa = 3.77VDD165 pKa = 4.95DD166 pKa = 4.75GSIKK170 pKa = 10.21HH171 pKa = 6.1ASSFVKK177 pKa = 10.43LQRR180 pKa = 11.84FDD182 pKa = 3.64PTSKK186 pKa = 8.93EE187 pKa = 3.98HH188 pKa = 4.85QVPRR192 pKa = 11.84VVSFRR197 pKa = 11.84QYY199 pKa = 10.59TNAFSLGRR207 pKa = 11.84YY208 pKa = 8.38IRR210 pKa = 11.84PLEE213 pKa = 4.12HH214 pKa = 6.42QFVNAPSFGRR224 pKa = 11.84FKK226 pKa = 10.82GYY228 pKa = 10.0RR229 pKa = 11.84WLAKK233 pKa = 9.13TRR235 pKa = 11.84NLTQRR240 pKa = 11.84ATDD243 pKa = 3.86ISLIWDD249 pKa = 4.44MIPNVCCVSIDD260 pKa = 3.35ASRR263 pKa = 11.84FEE265 pKa = 4.05AHH267 pKa = 6.03VSKK270 pKa = 11.29DD271 pKa = 3.35MLSLEE276 pKa = 4.17FEE278 pKa = 4.71TYY280 pKa = 10.39SDD282 pKa = 4.21AYY284 pKa = 10.69RR285 pKa = 11.84GDD287 pKa = 3.34ARR289 pKa = 11.84LVKK292 pKa = 9.6MLKK295 pKa = 10.69AMLATKK301 pKa = 8.92MHH303 pKa = 6.3TKK305 pKa = 10.63YY306 pKa = 10.78GLKK309 pKa = 10.32CSILGRR315 pKa = 11.84RR316 pKa = 11.84LSGDD320 pKa = 2.89MCTSLGNALLMCGMLNFISEE340 pKa = 4.56DD341 pKa = 3.46LNIDD345 pKa = 3.82FKK347 pKa = 11.18IYY349 pKa = 10.53DD350 pKa = 4.7DD351 pKa = 5.94GDD353 pKa = 3.91DD354 pKa = 4.88CLLFMNKK361 pKa = 9.36NLLPRR366 pKa = 11.84LEE368 pKa = 4.01EE369 pKa = 3.6MRR371 pKa = 11.84NCFKK375 pKa = 11.04RR376 pKa = 11.84LGFDD380 pKa = 3.27VRR382 pKa = 11.84VDD384 pKa = 4.05KK385 pKa = 10.83IAYY388 pKa = 8.35CLEE391 pKa = 4.44EE392 pKa = 4.24IVFCQSHH399 pKa = 7.07PIYY402 pKa = 9.29WAPGQGVLCPDD413 pKa = 4.17PDD415 pKa = 3.41KK416 pKa = 11.52HH417 pKa = 6.2MSALYY422 pKa = 9.79TGTSYY427 pKa = 11.14GRR429 pKa = 11.84SARR432 pKa = 11.84RR433 pKa = 11.84DD434 pKa = 3.34RR435 pKa = 11.84LIAFSTAMCCLSFLTNIPILGAFHH459 pKa = 7.05SMVHH463 pKa = 6.57RR464 pKa = 11.84ATATKK469 pKa = 10.42RR470 pKa = 11.84KK471 pKa = 9.06MIFPDD476 pKa = 3.07RR477 pKa = 11.84TLEE480 pKa = 3.78YY481 pKa = 10.12LYY483 pKa = 10.8KK484 pKa = 10.95NNSSCSKK491 pKa = 10.81CPGLNDD497 pKa = 3.62ALRR500 pKa = 11.84VSFYY504 pKa = 11.02LAFGYY509 pKa = 8.71TPSQQEE515 pKa = 3.66QLEE518 pKa = 4.34EE519 pKa = 4.38HH520 pKa = 6.93FDD522 pKa = 3.45TTLLHH527 pKa = 6.32FKK529 pKa = 10.31HH530 pKa = 6.0YY531 pKa = 11.07AEE533 pKa = 5.03LSQPVFDD540 pKa = 4.94YY541 pKa = 9.87ITWTSVTDD549 pKa = 3.94TPACQLDD556 pKa = 3.72APTT559 pKa = 4.45
MM1 pKa = 7.17FGYY4 pKa = 9.99RR5 pKa = 11.84RR6 pKa = 11.84HH7 pKa = 6.09ICSHH11 pKa = 6.6HH12 pKa = 6.69GNLHH16 pKa = 5.8TYY18 pKa = 7.89QRR20 pKa = 11.84EE21 pKa = 3.91RR22 pKa = 11.84SIVPCVVSSVCSSSSEE38 pKa = 3.9GAVIVGNKK46 pKa = 7.3WVAQPLGSGCSMSRR60 pKa = 11.84TNQQSEE66 pKa = 4.36SCSAKK71 pKa = 9.11FCKK74 pKa = 10.15FLTIKK79 pKa = 10.59NSRR82 pKa = 11.84TWDD85 pKa = 4.28PIVFSPCSHH94 pKa = 6.33NFMLAAKK101 pKa = 10.09SRR103 pKa = 11.84ILKK106 pKa = 7.52EE107 pKa = 4.22TPQPEE112 pKa = 4.09DD113 pKa = 3.29EE114 pKa = 4.31YY115 pKa = 11.47LRR117 pKa = 11.84RR118 pKa = 11.84FALSLHH124 pKa = 6.17AVAKK128 pKa = 10.47SIFVKK133 pKa = 8.71QTPLTRR139 pKa = 11.84EE140 pKa = 4.1EE141 pKa = 4.11FLSAYY146 pKa = 10.15SGSKK150 pKa = 9.11RR151 pKa = 11.84LRR153 pKa = 11.84YY154 pKa = 10.25AMAADD159 pKa = 3.66RR160 pKa = 11.84VYY162 pKa = 11.21DD163 pKa = 3.77VDD165 pKa = 4.95DD166 pKa = 4.75GSIKK170 pKa = 10.21HH171 pKa = 6.1ASSFVKK177 pKa = 10.43LQRR180 pKa = 11.84FDD182 pKa = 3.64PTSKK186 pKa = 8.93EE187 pKa = 3.98HH188 pKa = 4.85QVPRR192 pKa = 11.84VVSFRR197 pKa = 11.84QYY199 pKa = 10.59TNAFSLGRR207 pKa = 11.84YY208 pKa = 8.38IRR210 pKa = 11.84PLEE213 pKa = 4.12HH214 pKa = 6.42QFVNAPSFGRR224 pKa = 11.84FKK226 pKa = 10.82GYY228 pKa = 10.0RR229 pKa = 11.84WLAKK233 pKa = 9.13TRR235 pKa = 11.84NLTQRR240 pKa = 11.84ATDD243 pKa = 3.86ISLIWDD249 pKa = 4.44MIPNVCCVSIDD260 pKa = 3.35ASRR263 pKa = 11.84FEE265 pKa = 4.05AHH267 pKa = 6.03VSKK270 pKa = 11.29DD271 pKa = 3.35MLSLEE276 pKa = 4.17FEE278 pKa = 4.71TYY280 pKa = 10.39SDD282 pKa = 4.21AYY284 pKa = 10.69RR285 pKa = 11.84GDD287 pKa = 3.34ARR289 pKa = 11.84LVKK292 pKa = 9.6MLKK295 pKa = 10.69AMLATKK301 pKa = 8.92MHH303 pKa = 6.3TKK305 pKa = 10.63YY306 pKa = 10.78GLKK309 pKa = 10.32CSILGRR315 pKa = 11.84RR316 pKa = 11.84LSGDD320 pKa = 2.89MCTSLGNALLMCGMLNFISEE340 pKa = 4.56DD341 pKa = 3.46LNIDD345 pKa = 3.82FKK347 pKa = 11.18IYY349 pKa = 10.53DD350 pKa = 4.7DD351 pKa = 5.94GDD353 pKa = 3.91DD354 pKa = 4.88CLLFMNKK361 pKa = 9.36NLLPRR366 pKa = 11.84LEE368 pKa = 4.01EE369 pKa = 3.6MRR371 pKa = 11.84NCFKK375 pKa = 11.04RR376 pKa = 11.84LGFDD380 pKa = 3.27VRR382 pKa = 11.84VDD384 pKa = 4.05KK385 pKa = 10.83IAYY388 pKa = 8.35CLEE391 pKa = 4.44EE392 pKa = 4.24IVFCQSHH399 pKa = 7.07PIYY402 pKa = 9.29WAPGQGVLCPDD413 pKa = 4.17PDD415 pKa = 3.41KK416 pKa = 11.52HH417 pKa = 6.2MSALYY422 pKa = 9.79TGTSYY427 pKa = 11.14GRR429 pKa = 11.84SARR432 pKa = 11.84RR433 pKa = 11.84DD434 pKa = 3.34RR435 pKa = 11.84LIAFSTAMCCLSFLTNIPILGAFHH459 pKa = 7.05SMVHH463 pKa = 6.57RR464 pKa = 11.84ATATKK469 pKa = 10.42RR470 pKa = 11.84KK471 pKa = 9.06MIFPDD476 pKa = 3.07RR477 pKa = 11.84TLEE480 pKa = 3.78YY481 pKa = 10.12LYY483 pKa = 10.8KK484 pKa = 10.95NNSSCSKK491 pKa = 10.81CPGLNDD497 pKa = 3.62ALRR500 pKa = 11.84VSFYY504 pKa = 11.02LAFGYY509 pKa = 8.71TPSQQEE515 pKa = 3.66QLEE518 pKa = 4.34EE519 pKa = 4.38HH520 pKa = 6.93FDD522 pKa = 3.45TTLLHH527 pKa = 6.32FKK529 pKa = 10.31HH530 pKa = 6.0YY531 pKa = 11.07AEE533 pKa = 5.03LSQPVFDD540 pKa = 4.94YY541 pKa = 9.87ITWTSVTDD549 pKa = 3.94TPACQLDD556 pKa = 3.72APTT559 pKa = 4.45
Molecular weight: 63.7 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1392 |
559 |
833 |
696.0 |
78.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.543 ± 0.44 | 3.233 ± 0.415 |
5.46 ± 0.051 | 3.951 ± 0.096 |
5.819 ± 0.056 | 4.885 ± 0.138 |
2.73 ± 0.289 | 4.454 ± 0.011 |
4.813 ± 0.327 | 8.477 ± 0.593 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.802 ± 0.247 | 4.382 ± 0.581 |
5.029 ± 0.434 | 3.376 ± 0.092 |
6.537 ± 0.26 | 8.908 ± 0.444 |
5.675 ± 0.077 | 6.466 ± 0.755 |
1.078 ± 0.003 | 4.382 ± 0.158 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
